Cloning, sequencing, and characterization of a gene cluster involved in EDTA degradation from the bacterium BNC1
- PMID: 11157232
- PMCID: PMC92636
- DOI: 10.1128/AEM.67.2.688-695.2001
Cloning, sequencing, and characterization of a gene cluster involved in EDTA degradation from the bacterium BNC1
Abstract
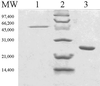
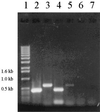
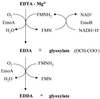
EDTA is a chelating agent, widely used in many industries. Because of its ability to mobilize heavy metals and radionuclides, it can be an environmental pollutant. The EDTA monooxygenases that initiate EDTA degradation have been purified and characterized in bacterial strains BNC1 and DSM 9103. However, the genes encoding the enzymes have not been reported. The EDTA monooxygenase gene was cloned by probing a genomic library of strain BNC1 with a probe generated from the N-terminal amino acid sequence of the monooxygenase. Sequencing of the cloned DNA fragment revealed a gene cluster containing eight genes. Two of the genes, emoA and emoB, were expressed in Escherichia coli, and the gene products, EmoA and EmoB, were purified and characterized. Both experimental data and sequence analysis showed that EmoA is a reduced flavin mononucleotide-utilizing monooxygenase and that EmoB is an NADH:flavin mononucleotide oxidoreductase. The two-enzyme system oxidized EDTA to ethylenediaminediacetate (EDDA) and nitrilotriacetate (NTA) to iminodiacetate (IDA) with the production of glyoxylate. The emoA and emoB genes were cotranscribed when BNC1 cells were grown on EDTA. Other genes in the cluster encoded a hypothetical transport system, a putative regulatory protein, and IDA oxidase that oxidizes IDA and EDDA. We concluded that this gene cluster is responsible for the initial steps of EDTA and NTA degradation.
Figures


References
-
- Alder A C, Siegrist H, Gujer W, Giger W. Behavior of NTA and EDTA in biological wastewater treatment. Water Res. 1990;24:733–742.
-
- Ausubel M F. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1993.
-
- Bergers P J M, DeGroot A C. The analysis of EDTA in water by HPLC. Water Res. 1994;28:639–642.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

