The adnA transcriptional factor affects persistence and spread of Pseudomonas fluorescens under natural field conditions
- PMID: 11157254
- PMCID: PMC92658
- DOI: 10.1128/AEM.67.2.852-857.2001
The adnA transcriptional factor affects persistence and spread of Pseudomonas fluorescens under natural field conditions
Abstract
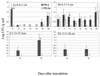
A soil plot was inoculated with a mixture of Pseudomonas fluorescens Pf0-2, the wild type, and Pf0-5a, a Tn5 insertion mutant in adnA, at 7.84 log CFU/g of soil. Over a period of 231 days, culturable populations of both strains were measured at selected times below and away from the point of inoculation. Pf0-5a did not spread as fast and attained significantly lower populations than Pf0-2. At sample depths below the inoculation site, the adnA mutant showed a significant decrease in CFU/g of soil as compared to Pf0-2. Pf0-2 was first detected at the 1.5-cm annular site at 3 days after inoculation, whereas Pf0-5a required 7 days to travel the same distance. At this distance, the wild-type strain could be detected at a 21.5- to 25-cm depth, whereas Pf0-5a could be detected only as deep as 15.5 to 18 cm. At 4.5 cm from the site of inoculation and in soil fractions corresponding to 13 to 18 cm, Pf0-2 was the only strain detected. These results suggest that the transcription factor AdnA provides a fitness advantage in P. fluorescens, allowing it to spread and survive in soil under field conditions.
Figures



Similar articles
-
Requirement of polyphosphate by Pseudomonas fluorescens Pf0-1 for competitive fitness and heat tolerance in laboratory media and sterile soil.Appl Environ Microbiol. 2009 Jun;75(12):3872-81. doi: 10.1128/AEM.00017-09. Epub 2009 Apr 24. Appl Environ Microbiol. 2009. PMID: 19395572 Free PMC article.
-
Transcriptional and antagonistic responses of Pseudomonas fluorescens Pf0-1 to phylogenetically different bacterial competitors.ISME J. 2011 Jun;5(6):973-85. doi: 10.1038/ismej.2010.196. Epub 2011 Jan 13. ISME J. 2011. PMID: 21228890 Free PMC article.
-
Colonization strategies of Pseudomonas fluorescens Pf0-1: activation of soil-specific genes important for diverse and specific environments.BMC Microbiol. 2013 Apr 27;13:92. doi: 10.1186/1471-2180-13-92. BMC Microbiol. 2013. PMID: 23622502 Free PMC article.
-
Use of in vivo expression technology to identify genes important in growth and survival of Pseudomonas fluorescens Pf0-1 in soil: discovery of expressed sequences with novel genetic organization.J Bacteriol. 2004 Nov;186(21):7411-9. doi: 10.1128/JB.186.21.7411-7419.2004. J Bacteriol. 2004. PMID: 15489453 Free PMC article.
-
Novel genes involved in Pseudomonas fluorescens Pf0-1 motility and biofilm formation.Appl Environ Microbiol. 2012 Jun;78(12):4318-29. doi: 10.1128/AEM.07201-11. Epub 2012 Apr 6. Appl Environ Microbiol. 2012. PMID: 22492452 Free PMC article.
Cited by
-
Pleiotropic effects of GacA on Pseudomonas fluorescens Pf0-1 in vitro and in soil.Appl Environ Microbiol. 2013 Sep;79(17):5405-10. doi: 10.1128/AEM.00819-13. Epub 2013 Jun 28. Appl Environ Microbiol. 2013. PMID: 23811507 Free PMC article.
-
Increased fitness of Pseudomonas fluorescens Pf0-1 leucine auxotrophs in soil.Appl Environ Microbiol. 2008 Jun;74(12):3644-51. doi: 10.1128/AEM.00429-08. Epub 2008 Apr 25. Appl Environ Microbiol. 2008. PMID: 18441116 Free PMC article.
-
Regulation of polyphosphate kinase production by antisense RNA in Pseudomonas fluorescens Pf0-1.Appl Environ Microbiol. 2012 Jun;78(12):4533-7. doi: 10.1128/AEM.07836-11. Epub 2012 Apr 6. Appl Environ Microbiol. 2012. PMID: 22492458 Free PMC article.
-
Alginate genes are required for optimal soil colonization and persistence by Pseudomonas fluorescens Pf0-1.Access Microbiol. 2019 May 3;1(3):e000021. doi: 10.1099/acmi.0.000021. eCollection 2019. Access Microbiol. 2019. PMID: 32974516 Free PMC article.
-
Requirement of polyphosphate by Pseudomonas fluorescens Pf0-1 for competitive fitness and heat tolerance in laboratory media and sterile soil.Appl Environ Microbiol. 2009 Jun;75(12):3872-81. doi: 10.1128/AEM.00017-09. Epub 2009 Apr 24. Appl Environ Microbiol. 2009. PMID: 19395572 Free PMC article.
References
-
- Amann R I. Fluorescently labeled, ribosomal-RNA-targeted oligonucleotide probes in the study of microbial ecology. Mol Ecol. 1995;4:543–553.
-
- Angle J S, Levin M A, McIntosh M, Glew J G. Pseudomonas aurofaciens in soil: survival and recovery efficiency. Microb Releases. 1994;2:247–254. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

