Genetic diversity of Pierce's disease strains and other pathotypes of Xylella fastidiosa
- PMID: 11157260
- PMCID: PMC92664
- DOI: 10.1128/AEM.67.2.895-903.2001
Genetic diversity of Pierce's disease strains and other pathotypes of Xylella fastidiosa
Abstract
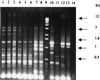
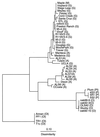
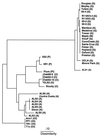
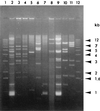
Strains of Xylella fastidiosa isolated from grape, almond, maple, and oleander were characterized by enterobacterial repetitive intergenic consensus sequence-, repetitive extragenic palindromic element (REP)-, and random amplified polymorphic DNA (RAPD)-PCR; contour-clamped homogeneous electric field (CHEF) gel electrophoresis; plasmid content; and sequencing of the 16S-23S rRNA spacer region. Combining methods gave greater resolution of strain groupings than any single method. Strains isolated from grape with Pierce's disease (PD) from California, Florida, and Georgia showed greater than previously reported genetic variability, including plasmid contents, but formed a cluster based on analysis of RAPD-PCR products, NotI and SpeI genomic DNA fingerprints, and 16S-23S rRNA spacer region sequence. Two groupings of almond leaf scorch (ALS) strains were distinguished by RAPD-PCR and CHEF gel electrophoresis, but some ALS isolates were clustered within the PD group. RAPD-PCR, CHEF gel electrophoresis, and 16S-23S rRNA sequence analysis produced the same groupings of strains, with RAPD-PCR resolving the greatest genetic differences. Oleander strains, phony peach disease (PP), and oak leaf scorch (OLS) strains were distinct from other strains. DNA profiles constructed by REP-PCR analysis were the same or very similar among all grape strains and most almond strains but different among some almond strains and all other strains tested. Eight of 12 ALS strains and 4 of 14 PD strains of X. fastidiosa isolated in California contained plasmids. All oleander strains carried the same-sized plasmid; all OLS strains carried the same-sized plasmid. A plum leaf scald strain contained three plasmids, two of which were the same sizes as those found in PP strains. These findings support a division of X. fastidiosa at the subspecies or pathovar level.
Figures


References
-
- Banks D, Albibi R, Chen J, Lamikanra O, Jarret R L, Smith B J. Specific detection of Xylella fastidiosa Pierce's disease strains. Curr Microbiol. 1999;39:85–88. - PubMed
-
- Barry T, Colleran G, Glennon M, Dunican L K, Gannon F. The 16S/23S ribosomal spacer region as a target for DNA probes to identify eubacteria. PCR Methods Appl. 1991;1:233–236. - PubMed
-
- Chang C J, Schaad N W. Electrophoretic protein profiles of total cell envelopes of xylem-limited plant pathogenic rickettsia-like bacteria (RLB) Phytopathology. 1982;72:935–936.
-
- Chen J, Chang C J, Jarret R L. Plasmids from Xylella fastidiosa. Can J Microbiol. 1992;38:993–995.
-
- Chen J, Chang C J, Jarret R L, Gawel N. Genetic variation among Xylella fastidiosa strains. Phytopathology. 1992;82:973–977.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

