Molecular analysis of Vibrio cholerae O1, O139, non-O1, and non-O139 strains: clonal relationships between clinical and environmental isolates
- PMID: 11157262
- PMCID: PMC92666
- DOI: 10.1128/AEM.67.2.910-921.2001
Molecular analysis of Vibrio cholerae O1, O139, non-O1, and non-O139 strains: clonal relationships between clinical and environmental isolates
Abstract
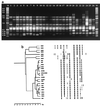
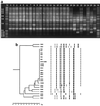
A total of 26 strains of Vibrio cholerae, including members of the O1, O139, and non-O1, non-O139 serogroups from both clinical and environmental sources, were examined for the presence of genes encoding cholera toxin (ctxA), zonula occludens toxin (zot), accessory cholera enterotoxin (ace), hemolysin (hlyA), NAG-specific heat-stable toxin (st), toxin-coregulated pilus (tcpA), and outer membrane protein (ompU), for genomic organization, and for the presence of the regulatory protein genes tcpI and toxR in order to determine relationships between epidemic serotypes and sources of isolation. While 22 of the 26 strains were hemolytic on 5% sheep blood nutrient agar, all strains were PCR positive for hlyA, the hemolysin gene. When multiplex PCR was used, all serogroup O1 and O139 strains were positive for tcpA, ompU, and tcpI. All O1 and O139 strains except one O1 strain and one O139 strain were positive for the ctxA, zot, and ace genes. Also, O1 strain VO3 was negative for the zot gene. All of the non-O1, non-O139 strains were negative for the ctxA, zot, ace, tcpA, and tcpI genes, and all of the non-O1, non-O139 strains except strain VO26 were negative for ompU. All of the strains except non-O1, non-O139 strain VO22 were PCR positive for the gene encoding the central regulatory protein, toxR. All V. cholerae strains were negative for the NAG-specific st gene. Of the nine non-ctx-producing strains of V. cholerae, only one, non-O1, non-O139 strain VO24, caused fluid accumulation in the rabbit ileal loop assay. The other eight strains, including an O1 strain, an O139 strain, and six non-O1, non-O139 strains, regardless of the source of isolation, caused fluid accumulation after two to five serial passages through the rabbit gut. Culture filtrates of all non-cholera-toxigenic strains grown in AKI media also caused fluid accumulation, suggesting that a new toxin was produced in AKI medium by these strains. Studies of clonality performed by using enterobacterial repetitive intergenic consensus sequence PCR, Box element PCR, amplified fragment length polymorphism (AFLP), and pulsed-field gel electrophoresis (PFGE) collectively indicated that the V. cholerae O1 and O139 strains had a clonal origin, whereas the non-O1, non-O139 strains belonged to different clones. The clinical isolates closely resembled environmental isolates in their genomic patterns. Overall, there was an excellent correlation among the results of the PCR, AFLP, and PFGE analyses, and individual strains derived from clinical and environmental sources produced similar fingerprint patterns. From the results of this study, we concluded that the non-cholera-toxin-producing strains of V. cholerae, whether of clinical or environmental origin, possess the ability to produce a new secretogenic toxin that is entirely different from the toxin produced by toxigenic V. cholerae O1 and O139 strains. We also concluded that the aquatic environment is a reservoir for V. cholerae O1, O139, non-O1, and non-O139 serogroup strains.
Figures


References
-
- Albert M J, Siddique A K, Islam M S, Faruque A S G, Ansaruzzaman M, Faruque S M, Sack R B. Large outbreak of clinical cholera due to Vibrio cholerae non-O1 in Bangladesh. Lancet. 1993;341:704. - PubMed
-
- Alm R A, Mayerhofer G, Kotlarski I, Manning P A. Amino-terminal domain of the El Tor hemolysin of Vibrio cholerae O1 is expressed in classical strain and is cytotoxic. Vaccine. 1991;9:588–594. - PubMed
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Short protocols in molecular biology. 3rd ed. New York, N.Y: John Wiley and Sons Inc.; 1995.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

