Portable system for microbial sample preparation and oligonucleotide microarray analysis
- PMID: 11157263
- PMCID: PMC92667
- DOI: 10.1128/AEM.67.2.922-928.2001
Portable system for microbial sample preparation and oligonucleotide microarray analysis
Abstract
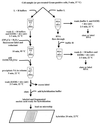
We have developed a three-component system for microbial identification that consists of (i) a universal syringe-operated silica minicolumn for successive DNA and RNA isolation, fractionation, fragmentation, fluorescent labeling, and removal of excess free label and short oligonucleotides; (ii) microarrays of immobilized oligonucleotide probes for 16S rRNA identification; and (iii) a portable battery-powered device for imaging the hybridization of fluorescently labeled RNA fragments with the arrays. The minicolumn combines a guanidine thiocyanate method of nucleic acid isolation with a newly developed hydroxyl radical-based technique for DNA and RNA labeling and fragmentation. DNA and RNA can also be fractionated through differential binding of double- and single-stranded forms of nucleic acids to the silica. The procedure involves sequential washing of the column with different solutions. No vacuum filtration steps, phenol extraction, or centrifugation is required. After hybridization, the overall fluorescence pattern is captured as a digital image or as a Polaroid photo. This three-component system was used to discriminate Escherichia coli, Bacillus subtilis, Bacillus thuringiensis, and human HL60 cells. The procedure is rapid: beginning with whole cells, it takes approximately 25 min to obtain labeled DNA and RNA samples and an additional 25 min to hybridize and acquire the microarray image using a stationary image analysis system or the portable imager.
Figures



References
-
- Barsky Y, Grammatin A, Ivanov A, Kreindlin E, Kotova E, Barskii V, Mirzabekov A. Wide-field luminescence microscopes for analyzing biological microchips. J Opt Technol. 1998;65:938–941.
-
- Bowtell D D L. Options available—from start to finish—for obtaining expression data by microarray. Nat Genet. 1999;21:25–32. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

