Impairment of lagging strand synthesis triggers the formation of a RuvABC substrate at replication forks
- PMID: 11157768
- PMCID: PMC133471
- DOI: 10.1093/emboj/20.3.619
Impairment of lagging strand synthesis triggers the formation of a RuvABC substrate at replication forks
Abstract
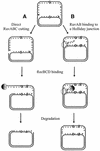
The holD gene codes for the psi subunit of the Escherichia coli DNA polymerase III holoenzyme, a component of the gamma complex clamp loader. A holD mutant was isolated for the first time in a screen for mutations that increase the frequency of tandem repeat deletions. In contrast to tandem repeat deletions in wild-type strains, deletion events stimulated by the holD mutation require RecA. They do not require RecF, and hence do not result from the recombinational repair of gaps, arguing against uncoupling of the leading and lagging strand polymerases in the holD mutant. The holD recBC combination of mutations is lethal and holD recBts recCts strains suffer DNA double-strand breaks (DSBs) at restrictive temperature. DSBs require the presence of the Holliday junction-specific enzymes RuvABC and are prevented in the presence of RecBCD. We propose that impairment of replication due to the holD mutation causes the arrest of the entire replisome; consequently, Holliday junctions are formed by replication fork reversal, and unequal crossing over during RecA- and RecBCD-mediated re-incorporation of reversed forks causes the hyper-recombination phenotype.
Figures


References
-
- Anderson D.G. and Kowalczykowski,S.C. (1997) The translocating RecBCD enzyme stimulates recombination by directing RecA protein onto ssDNA in a χ-regulated manner. Cell, 90, 77–86. - PubMed
-
- Ashley C.T.J. and Warren,S.T. (1995) Trinucleotide repeat expansion and human disease. Annu. Rev. Genet., 29, 703–728. - PubMed
-
- Ason B., Bertram,J.G., Hingorani,M.M., Beechem,J.M., O’Donnell,M., Goodman,M.F. and Bloom,L.B. (2000) A model for Escherichia coli DNA polymerase III holoenzyme assembly at primer/template ends—DNA triggers a change in binding specificity of the γ complex clamp loader. J. Biol. Chem., 275, 3006–3015. - PubMed
-
- Bierne H., Seigneur,M., Ehrlich,S.D. and Michel,B. (1997a) uvrD mutations enhance tandem repeat deletion in the Escherichia coli chromosome via SOS induction of the RecF recombination pathway. Mol. Microbiol., 26, 557–567. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

