The retinoic acid-metabolizing enzyme, CYP26A1, is essential for normal hindbrain patterning, vertebral identity, and development of posterior structures
- PMID: 11157778
- PMCID: PMC312609
- DOI: 10.1101/gad.855001
The retinoic acid-metabolizing enzyme, CYP26A1, is essential for normal hindbrain patterning, vertebral identity, and development of posterior structures
Abstract
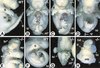
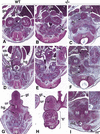
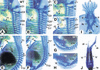
The active derivative of vitamin A, retinoic acid (RA), is essential for normal embryonic development. The spatio-temporal distribution of embryonic RA results from regulated expression of RA-synthesizing retinaldehyde dehydrogenases and RA-metabolizing cytochrome P450s (CYP26). Excess RA administration or RA deficiency results in a complex spectrum of embryonic abnormalities. As a first step in understanding the developmental function of RA-metabolizing enzymes, we have disrupted the murine Cyp26A1 gene. We report that Cyp26A1-null mutants die during mid-late gestation and show a number of major morphogenetic defects. Spina bifida and truncation of the tail and lumbosacral region (including abnormalities of the kidneys, urogenital tract, and hindgut) are the most conspicuous defects, leading in extreme cases to a sirenomelia ("mermaid tail") phenotype. Cyp26A1 mutants also show posterior transformations of cervical vertebrae and abnormal patterning of the rostral hindbrain, which appears to be partially posteriorly transformed. These defects correlate with two major sites of Cyp26A1 expression in the rostral neural plate and embryonic tail bud. Because all of the Cyp26A1(-/-) abnormalities closely resemble RA teratogenic effects, we postulate that the key function of CYP26A1 is to maintain specific embryonic areas in a RA-depleted state, to protect them against the deleterious effect of ectopic RA signaling.
Figures




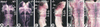


References
-
- Abu-Abed SS, Beckett BR, Chiba H, Chithalen JV, Jones G, Metzger D, Chambon P, Petkovich M. Mouse P450RAI (CYP26) expression and retinoic acid-inducible retinoic acid metabolism in F9 cells are regulated by retinoic acid receptor γ and retinoid X receptor α. J Biol Chem. 1998;273:2409–2415. - PubMed
-
- Burke AC, Nelson CE, Morgan BA, Tabin C. Hox genes and the evolution of vertebrate axial morphology. Development. 1995;121:333–346. - PubMed
-
- Chambon P. A decade of molecular biology of retinoic acid receptors. FASEB J. 1996;10:940–954. - PubMed
-
- Chazaud C, Chambon P, Dollé P. Retinoic acid is required in the mouse embryo for left-right asymmetry determination and heart morphogenesis. Development. 1999;126:2589–2596. - PubMed
-
- Conlon RA. Retinoic acid and pattern formation in vertebrates. Trends Genet. 1995;11:314–319. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
