Low-complexity regions in Plasmodium falciparum proteins
- PMID: 11157785
- PMCID: PMC311019
- DOI: 10.1101/gr.gr-1522r
Low-complexity regions in Plasmodium falciparum proteins
Abstract
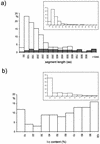
Full-sequence data available for Plasmodium falciparum chromosomes 2 and 3 are exploited to perform a statistical analysis of the long tracts of biased amino acid composition that characterize the vast majority of P. falciparum proteins and to make a comparison with similarly defined tracts from other simple eukaryotes. When the relatively minor subset of prevalently hydrophobic segments is discarded from the set of low-complexity segments identified by current segmentation methods in P. falciparum proteins, a good correspondence is found between prevalently hydrophilic low-complexity segments and the species-specific, rapidly diverging insertions detected by multiple-alignment procedures when sequences of bona fide homologs are available. Amino acid preferences are fairly uniform in the set of hydrophilic low-complexity segments identified in the two P. falciparum chromosomes sequenced, as well as in sequenced genes from Plasmodium berghei, but differ from those observed in Saccharomyces cerevisiae and Dictyostelium discoideum. In the two plasmodial species, amino acid frequencies do not correlate with properties such as hydrophilicity, small volume, or flexibility, which might be expected to characterize residues involved in nonglobular domains but do correlate with A-richness in codons. An effect of phenotypic selection versus neutral drift, however, is suggested by the predominance of asparagine over lysine.
Figures



Comment in
-
Low-complexity regions in Plasmodium proteins: in search of a function.Genome Res. 2001 Feb;11(2):195-7. doi: 10.1101/gr.176401. Genome Res. 2001. PMID: 11157782 No abstract available.
References
-
- Bell SJ, Forsdyke DR. Deviations from Chargaff's second parity rule correlate with direction of transcription. J Theor Biol. 1999;197:63–76. - PubMed
-
- Birago C, Pace T, Picci L, Pizzi E, Scotti R, Ponzi M. The putative gene for the first enzyme of glutathione biosynthesis in P. berghei and P. falciparum. Mol Biochem Parasitol. 1999;99:33–40. - PubMed
-
- Bowman S, Lawson D, Basham D, Brown D, Chillingworth T, Churcher CM, Craig A, Davies RM, Devlin K, Feltwell T, et al. The complete nucleotide sequence of chromosome 3 of P. falciparum. Nature. 1999;400:532–538. - PubMed
-
- Braun JV, Mueller HG. Statistical methods for DNA sequence segmentation. Statist Sci. 1998;13:142–162.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
