New family of regulators in the environmental signaling pathway which activates the general stress transcription factor sigma(B) of Bacillus subtilis
- PMID: 11157946
- PMCID: PMC95007
- DOI: 10.1128/JB.183.4.1329-1338.2001
New family of regulators in the environmental signaling pathway which activates the general stress transcription factor sigma(B) of Bacillus subtilis
Abstract
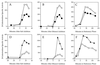
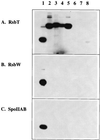
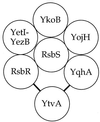
Expression of the general stress regulon of Bacillus subtilis is controlled by the alternative transcription factor sigma(B), which is activated when cells encounter growth-limiting energy or environmental stresses. The RsbT serine-threonine kinase is required to convey environmental stress signals to sigma(B), and this kinase activity is magnified in vitro by the RsbR protein, a positive regulator important for full in vivo response to salt or heat stress. Previous genetic analysis suggested that RsbR function is redundant with other unidentified regulators. A search of the translated B. subtilis genome found six paralogous proteins with significant similarity to RsbR: YetI, YezB, YkoB, YojH, YqhA, and YtvA. Their possible regulatory roles were investigated using three different approaches. First, genetic analysis found that null mutations in four of the six paralogous genes have marked effects on the sigma(B) environmental signaling pathway, either singly or in combination. The two exceptions were yetI and yezB, adjacent genes which appear to encode a split paralog. Second, biochemical analysis found that YkoB, YojH, and YqhA are specifically phosphorylated in vitro by the RsbT environmental signaling kinase, as had been previously shown for RsbR, which is phosphorylated on two threonine residues in its C-terminal region. Both residues are conserved in the three phosphorylated paralogs but are absent in the ones that were not substrates of RsbT: YetI and YezB, each of which bears only one of the conserved residues; and YtvA, which lacks both residues and instead possesses an N-terminal PAS domain. Third, analysis in the yeast two-hybrid system suggested that all six paralogs interact with each other and with the RsbR and RsbS environmental regulators. Our data indicate that (i) RsbR, YkoB, YojH, YqhA, and YtvA function in the environmental stress signaling pathway; (ii) YtvA acts as a positive regulator; and (iii) RsbR, YkoB, YojH, and YqhA collectively act as potent negative regulators whose loss increases sigma(B) activity more than 400-fold in unstressed cells.
Figures
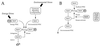



References
-
- Akbar S, Kang C M, Gaidenko T A, Price C W. Modulator protein RsbR regulates environmental signalling in the general stress pathway of Bacillus subtilis. Mol Microbiol. 1997;24:567–578. - PubMed
-
- Alper S, Dufour A, Garsin D A, Duncan L, Losick R. Role of adenosine nucleotides in the regulation of a stress-response transcription factor in Bacillus subtilis. J Mol Biol. 1996;260:165–177. - PubMed
-
- Alper S, Duncan L, Losick R. An adenosine nucleotide switch controlling the activity of a cell type-specific transcription factor in B. subtilis. Cell. 1994;77:195–205. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

