Drosophila as a model host for Pseudomonas aeruginosa infection
- PMID: 11157963
- PMCID: PMC95024
- DOI: 10.1128/JB.183.4.1466-1471.2001
Drosophila as a model host for Pseudomonas aeruginosa infection
Abstract
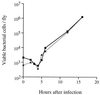
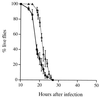
Using the fruit fly Drosophila melanogaster as model host, we have identified mutants of the bacterium Pseudomonas aeruginosa with reduced virulence. Strikingly, all strains strongly impaired in fly killing also lacked twitching motility; most such strains had a mutation in pilGHIJKL chpABCDE, a gene cluster known to be required for twitching motility and potentially encoding a signal transduction system. The pil chp genes appear to control the expression of additional virulence factors, however, since the wild-type fly-killing phenotype of a subset of mutants isolated on the basis of their compact colony morphology indicated that twitching motility itself was not required for full virulence in the fly.
Figures



References
-
- Alm R A, Mattick J S. Genes involved in the biogenesis and function of type-4 fimbriae in Pseudomonas aeruginosa. Gene. 1997;192:89–98. - PubMed
-
- Anderson K V. Toll signaling pathways in the innate immune response. Curr Opin Immunol. 2000;12:13–19. - PubMed
-
- Boman H G. Antibacterial peptides: key components needed in immunity. Cell. 1991;65:205–207. - PubMed
-
- Boman H G, Nilsson I, Rasmuson B. Inducible antibacterial defence system in Drosophila. Nature. 1972;237:232–235. - PubMed
-
- Bradley D E. A function of Pseudomonas aeruginosa PAO polar pili: twitching motility. Can J Microbiol. 1980;26:146–160. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

