Identification and characterization of an escorter for two secretory adhesins in Toxoplasma gondii
- PMID: 11157983
- PMCID: PMC2196004
- DOI: 10.1083/jcb.152.3.563
Identification and characterization of an escorter for two secretory adhesins in Toxoplasma gondii
Abstract
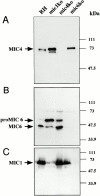
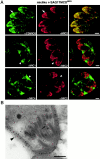
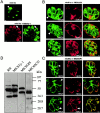
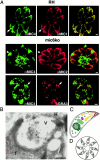
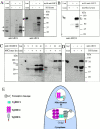
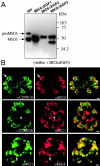
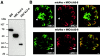
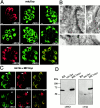
The intracellular protozoan parasite Toxoplasma gondii shares with other members of the Apicomplexa a common set of apical structures involved in host cell invasion. Micronemes are apical secretory organelles releasing their contents upon contact with host cells. We have identified a transmembrane micronemal protein MIC6, which functions as an escorter for the accurate targeting of two soluble proteins MIC1 and MIC4 to the micronemes. Disruption of MIC1, MIC4, and MIC6 genes allowed us to precisely dissect their contribution in sorting processes. We have mapped domains on these proteins that determine complex formation and targeting to the organelle. MIC6 carries a sorting signal(s) in its cytoplasmic tail whereas its association with MIC1 involves a lumenal EGF-like domain. MIC4 binds directly to MIC1 and behaves as a passive cargo molecule. In contrast, MIC1 is linked to a quality control system and is absolutely required for the complex to leave the early compartments of the secretory pathway. MIC1 and MIC4 bind to host cells, and the existence of such a complex provides a plausible mechanism explaining how soluble adhesins act. We hypothesize that during invasion, MIC6 along with adhesins establishes a bridge between the host cell and the parasite.
Figures

References
-
- Achbarou A., Mercereau-Puijalon O., Autheman J.M., Fortier B., Camus D., Dubremetz J.F. Characterization of microneme proteins of Toxoplasma gondii . Mol. Biochem. Parasitol. 1991;47:223–233. - PubMed
-
- Barnwell J.W., Galinski M.R. Plasmodium vivaxa glimpse into the unique and shared biology of the merozoite. Ann. Trop. Med. Parasitol. 1995;89:113–120. - PubMed
-
- Bastin P., Bagherzadeh Z., Matthews K.R., Gull K. A novel epitope tag system to study protein targeting and organelle biogenesis in Trypanosoma brucei . Mol. Biochem. Parasitol. 1996;77:235–239. - PubMed
-
- Bermudes D., Dubremetz J.F., Achbarou A., Joiner K.A. Cloning of a cDNA encoding the dense granule protein GRA3 from Toxoplasma gondii . Mol. Biochem. Parasitol. 1994;68:247–257. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

