Rapid typing of human adenoviruses by a general PCR combined with restriction endonuclease analysis
- PMID: 11158096
- PMCID: PMC87765
- DOI: 10.1128/JCM.39.2.498-505.2001
Rapid typing of human adenoviruses by a general PCR combined with restriction endonuclease analysis
Abstract
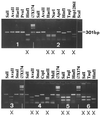
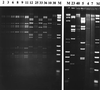
We have developed a system for rapid typing of adenoviruses (Ads) based on a combination of PCR and restriction endonuclease (RE) digestion (PCR-RE digestion). Degenerated consensus primers were designed, allowing amplification of DNA from all 51 human Ad prototype strains and altogether 44 different genome variants of Ad serotypes 1, 3, 4, 5, 7, 11, 19, 40, and 41. The 301-bp amplimer of 22 prototype strains representing all six subgenera and the genome variant was selected as a target for sequencing to look for subgenus and genome type variabilities. The sequences obtained were used to facilitate the selection of specific REs for discrimination purposes in a diagnostic assay by following the concept of cleavage or noncleavage of the 301-bp amplimer. On the basis of these results, a flowchart was constructed, allowing identification of subgenus B:2 and D serotypes and almost complete distinction of subgenus A, B:1, C, E, and F serotypes. Application of the PCR-RE digestion system to clinical samples allowed typing of 34 of 40 clinical samples positive for Ad. The genome type determined by this method was identical to that obtained by traditional RE typing of full-length Ad DNA. The remaining six samples were positive only after a nested PCR. Therefore, to reduce the risk of false-negative results, samples scored negative by the PCR-RE digestion system should be evaluated by the described nested PCR. Used in combination, the PCR-RE digestion method and the nested PCR provide a reliable and sensitive system that can easily be applied to all kinds of clinical samples when rapid identification of adenoviruses is needed.
Figures




References
-
- Adrian T, Sassinek J, Wigand R. Genome type analysis of 480 isolates of adenovirus types 1, 2, and 5. Arch Virol. 1990;112:235–248. - PubMed
-
- Adrian T H, Wadell G, Hierholzer J C, Wigand R. DNA restriction analysis of adenovirus prototypes 1 to 41. Arch Virol. 1986;91:277–290. - PubMed
-
- Akusjärvi G, Aleström P, Pettersson M, Lager M, Jörnvall H, Pettersson U. The gene for the adenovirus 2 hexon polypeptide. J Biol Chem. 1984;259:13976–13979. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

