The Chinese hamster dihydrofolate reductase replication origin beta is active at multiple ectopic chromosomal locations and requires specific DNA sequence elements for activity
- PMID: 11158297
- PMCID: PMC99564
- DOI: 10.1128/MCB.21.4.1098-1110.2001
The Chinese hamster dihydrofolate reductase replication origin beta is active at multiple ectopic chromosomal locations and requires specific DNA sequence elements for activity
Erratum in
- Mol Cell Biol 2002 Sep;22(17):6319
Abstract
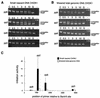
To identify cis-acting genetic elements essential for mammalian chromosomal DNA replication, a 5.8-kb fragment from the Chinese hamster dihydrofolate reductase (DHFR) locus containing the origin beta (ori-beta) initiation region was stably transfected into random ectopic chromosomal locations in a hamster cell line lacking the endogenous DHFR locus. Initiation at ectopic ori-beta in uncloned pools of transfected cells was measured using a competitive PCR-based nascent strand abundance assay and shown to mimic that at the endogenous ori-beta region in Chinese hamster ovary K1 cells. Initiation activity of three ectopic ori-beta deletion mutants was reduced, while the activity of another deletion mutant was enhanced. The results suggest that a 5.8-kb fragment of the DHFR ori-beta region is sufficient to direct initiation and that specific DNA sequences in the ori-beta region are required for efficient initiation activity.
Figures



References
-
- Abdurashidova G, Deganuto M, Klima R, Riva S, Biamonti G, Giacca M, Falaschi A. Start sites of bidirectional DNA synthesis at the human lamin B2 origin. Science. 2000;287:2023–2026. - PubMed
-
- Aladjem M I, Groudine M, Brody L L, Dieken E S, Fournier K, Wahl G M, Epner E M. Participation of the human β-globin locus control region in initiation of DNA replication. Science. 1995;270:815–819. - PubMed
-
- Aladjem M I, Rodewald L W, Koleman J L, Wahl G M. Genetic dissection of a mammalian replicator in the human β-globin locus. Science. 1998;281:1005–1009. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
