Human L1 retrotransposition: cis preference versus trans complementation
- PMID: 11158327
- PMCID: PMC99594
- DOI: 10.1128/MCB.21.4.1429-1439.2001
Human L1 retrotransposition: cis preference versus trans complementation
Abstract
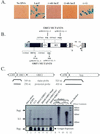
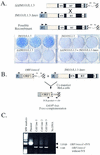
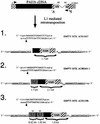
Long interspersed nuclear elements (LINEs or L1s) comprise approximately 17% of human DNA; however, only about 60 of the approximately 400,000 L1s are mobile. Using a retrotransposition assay in cultured human cells, we demonstrate that L1-encoded proteins predominantly mobilize the RNA that encodes them. At much lower levels, L1-encoded proteins can act in trans to promote retrotransposition of mutant L1s and other cellular mRNAs, creating processed pseudogenes. Mutant L1 RNAs are mobilized at 0.2 to 0.9% of the retrotransposition frequency of wild-type L1s, whereas cellular RNAs are mobilized at much lower frequencies (ca. 0.01 to 0.05% of wild-type levels). Thus, we conclude that L1-encoded proteins demonstrate a profound cis preference for their encoding RNA. This mechanism could enable L1 to remain retrotransposition competent in the presence of the overwhelming number of nonfunctional L1s present in human DNA.
Figures


References
-
- Boeke J D. LINEs and Alus—the polyA connection. Nat Genet. 1997;16:6–7. - PubMed
-
- Boeke J D, Stoye J P. Retrotransposons, endogenous retroviruses, and the evolution of retroelements. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 343–435. - PubMed
-
- Boissinot S, Chevret P, Furano A V. L1 (LINE-1) retrotransposon evolution and amplification in recent human history. Mol Biol Evol. 2000;17:915–928. - PubMed
-
- Cost G J, Boeke J D. Targeting of human retrotransposon integration is directed by the specificity of the L1 endonuclease for regions of unusual DNA structure. Biochemistry. 1998;37:18081–18093. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
