CD8+ T cells recognize an inclusion membrane-associated protein from the vacuolar pathogen Chlamydia trachomatis
- PMID: 11158611
- PMCID: PMC14725
- DOI: 10.1073/pnas.98.3.1160
CD8+ T cells recognize an inclusion membrane-associated protein from the vacuolar pathogen Chlamydia trachomatis
Abstract
During infection with Chlamydia trachomatis, CD8(+) T cells are primed, even though the bacteria remain confined to a host cell vacuole throughout their developmental cycle. Because CD8(+) T cells recognize antigens processed from cytosolic proteins, the Chlamydia antigens recognized by these CD8(+) T cells very likely have access to the host cell cytoplasm during infection. The identity of these C. trachomatis proteins has remained elusive, even though their localization suggests they may play important roles in the biology of the organism. Here we use a retroviral expression system to identify Cap1, a 31-kDa protein from C. trachomatis recognized by protective CD8(+) T cells. Cap1 contains no strong homology to any known protein. Immunofluorescence microscopy by using Cap1-specific antibody demonstrates that this protein is localized to the vacuolar membrane. Cap1 is virtually identical among the human C. trachomatis serovars, suggesting that a vaccine incorporating Cap1 might enable the vaccine to protect against all C. trachomatis serovars. The identification of proteins such as Cap1 that associate with the inclusion membrane will be required to fully understand the interaction of C. trachomatis with its host cell.
Figures

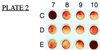
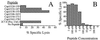
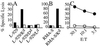
 )
or uninfected
(
)
or uninfected
( )
Balb/3T3 cells.
)
Balb/3T3 cells.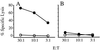
 )
or untreated
(
)
or untreated
( )
P815 cells. Graphs are representative of three experimental and three
control mice.
)
P815 cells. Graphs are representative of three experimental and three
control mice.
References
-
- Burstein G R, Gaydos C A, Diener-West M, Howell M R, Zenilman J M, Quinn T C. J Am Med Assoc. 1998;280:521–526. - PubMed
-
- Ward M E. In: Chlamydia: Intracellular Biology, Pathogenesis, and Immunity. Stephens R S, editor. Washington, DC: Am. Soc. Microbiol.; 1999. pp. 171–210.
-
- Shor A, Phillips J I. J Am Med Assoc. 1999;282:2071–2073. - PubMed
-
- Igietseme J U, Ramsey K H, Magee D M, Williams D M, Kincy T J, Rank R G. Reg Immunol. 1993;5:317–324. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

