Inhibition of cytochrome P450 (CYP450) isoforms by isoniazid: potent inhibition of CYP2C19 and CYP3A
- PMID: 11158730
- PMCID: PMC90302
- DOI: 10.1128/AAC.45.2.382-392.2001
Inhibition of cytochrome P450 (CYP450) isoforms by isoniazid: potent inhibition of CYP2C19 and CYP3A
Abstract
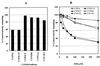
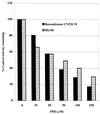
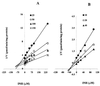
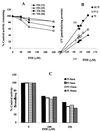
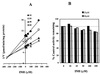
Isoniazid (INH) remains the most safe and cost-effective drug for the treatment and prophylaxis of tuberculosis. The use of INH has increased over the past years, largely as a result of the coepidemic of human immunodeficiency virus infection. It is frequently given chronically to critically ill patients who are coprescribed multiple medications. The ability of INH to elevate the concentrations in plasma and/or toxicity of coadministered drugs, including those of narrow therapeutic range (e.g., phenytoin), has been documented in humans, but the mechanisms involved are not well understood. Using human liver microsomes (HLMs), we tested the inhibitory effect of INH on the activity of common drug-metabolizing human cytochrome P450 (CYP450) isoforms using isoform-specific substrate probe reactions. Incubation experiments were performed at a single concentration of each substrate probe at its K(m) value with a range of INH concentrations. CYP2C19 and CYP3A were inhibited potently by INH in a concentration-dependent manner. At 50 microM INH (approximately 6.86 microg/ml), the activities of these isoforms decreased by approximately 40%. INH did not show significant inhibition (<10% at 50 microM) of other isoforms (CYP2C9, CYP1A2, and CYP2D6). To accurately estimate the inhibition constants (K(i) values) for each isoform, four concentrations of INH were incubated across a range of five concentrations of specific substrate probes. The mean K(i) values (+/- standard deviation) for the inhibition of CYP2C19 by INH in HLMs and recombinant human CYP2C19 were 25.4 +/- 6.2 and 13 +/- 2.4 microM, respectively. INH showed potent noncompetitive inhibition of CYP3A (K(i) = 51.8 +/- 2.5 to 75.9 +/- 7.8 microM, depending on the substrate used). INH was a weak noncompetitive inhibitor of CYP2E1 (K(i) = 110 +/- 33 microM) and a competitive inhibitor of CYP2D6 (K(i) = 126 +/- 23 microM), but the mean K(i) values for the inhibition of CYP2C9 and CYP1A2 were above 500 microM. Inhibition of one or both CYP2C19 and CYP3A isoforms is the likely mechanism by which INH slows the elimination of coadministered drugs, including phenytoin, carbamazepine, diazepam, triazolam, and primidone. Slow acetylators of INH may be at greater risk for adverse drug interactions, as the degree of inhibition was concentration dependent. These data provide a rational basis for understanding drug interaction with INH and predict that other drugs metabolized by these two enzymes may also interact.
Figures
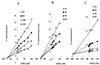

References
-
- Bajpai M, Roskos L K, Shen D D, Levy R H. Roles of cytochrome P4502C9 and cytochrome P4502C19 in the stereoselective metabolism of phenytoin to its major metabolite. Drug Metab Dispos. 1996;24:1401–1403. - PubMed
-
- Bertz R J, Granneman G R. Use of in vitro and in vivo data to estimate the likelihood of metabolic pharmacokinetic interactions. Clin Pharmacokinet. 1997;32:210–258. - PubMed
-
- Brennan R W, Dehejia H, Kutt H, Verebely K, McDowell F. Diphenylhydantoin intoxication attendant to slow inactivation of isoniazid. Neurology. 1970;20:687–693. - PubMed
-
- Brodie M J, Boobis A R, Hillyard C J, Abeyasekera G, MacIntyre I, Park B K. Effect of isoniazid on vitamin D metabolism and hepatic monooxygenase activity. Clin Pharmacol Ther. 1981;30:363–367. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

