Transcriptional regulation of the Drosophila gene zen by competing Smad and Brinker inputs
- PMID: 11159914
- PMCID: PMC312624
- DOI: 10.1101/gad.861401
Transcriptional regulation of the Drosophila gene zen by competing Smad and Brinker inputs
Abstract
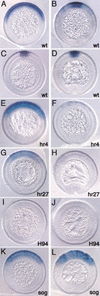
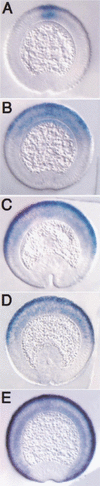
The establishment of expression domains of developmentally regulated genes depends on cues provided by different concentrations of transcriptional activators and repressors. Here we analyze the regulation of the Drosophila gene zen, which is a target of the Decapentaplegic (Dpp) signaling pathway during cellular blastoderm formation. We show that low levels of the Dpp signal transducer p-Mad (phosphorylated Mad), together with the recently discovered negative regulator Brinker (Brk), define the spatial limits of zen transcription in a broad dorsal-on/ventral-off domain. The subsequent refinement of this pattern to the dorsal-most cells, however, correlates with high levels of p-Mad that accumulate in the same region during late blastoderm. Examination of the zen regulatory sequences revealed the presence of multiple Mad and Brk binding sites, and our results indicate that a full occupancy of the Mad sites due to high concentrations of nuclear Mad is the primary mechanism for refinement of zen. Interestingly, several Mad and Brk binding sites overlap, and we show that Mad and Brk cannot bind simultaneously to such sites. We propose a model whereby competition between Mad and Brk determines spatially restricted domains of expression of Dpp target genes.
Figures




References
-
- Ashe HL, Levine M. Local inhibition and long-range enhancement of Dpp signal transduction by Sog. Nature. 1999;398:427–431. - PubMed
-
- Ashe HL, Mannervik M, Levine M. Dpp signaling thresholds in the dorsal ectoderm of the Drosophila embryo. Development. 2000;127:3305–3312. - PubMed
-
- Biehs B, François V, Bier E. The Drosophila short gastrulation gene prevents Dpp from autoactivating and suppressing neurogenesis in the neuroectoderm. Genes & Dev. 1996;10:2922–2934. - PubMed
-
- Campbell G, Tomlinson A. Transducing the Dpp morphogen gradient in the wing of Drosophila: Regulation of Dpp target genes by brinker. Cell. 1999;96:553–562. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
