Identification of Rgg-regulated exoproteins of Streptococcus pyogenes
- PMID: 11159974
- PMCID: PMC97958
- DOI: 10.1128/IAI.69.2.822-831.2001
Identification of Rgg-regulated exoproteins of Streptococcus pyogenes
Abstract
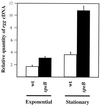
Streptococcus pyogenes secretes many proteins that influence host-pathogen interactions. Despite their importance, relatively little is known about the regulation of these proteins. The rgg gene (also known as ropB) is required for the expression of streptococcal erythrogenic toxin B (SPE B), an extracellular cysteine protease that contributes to virulence. Proteomics was used to determine if rgg regulates the expression of additional exoproteins. Exponential- and stationary-phase culture supernatant proteins made by S. pyogenes NZ131 rgg and NZ131 speB were separated by two-dimensional electrophoresis. Differences were identified in supernatant proteins from both exponential- and stationary-phase cultures, although considerably more differences were detected among stationary-phase supernatant proteins. Forty-two proteins were identified by peptide fingerprinting with matrix-assisted laser desorption mass spectrometry. Mitogenic factor, DNA entry nuclease (open reading frame [ORF 226]), and ORF 953, which has no known function, were more abundant in the culture supernatants of the rgg mutant compared to the speB mutant. ClpB, lysozyme, and autolysin were detected in the culture supernatant of the speB mutant but not the rgg mutant. To determine if Rgg affected protein expression at the transcriptional level, real-time (TaqMan) reverse transcription (RT)-PCR was used to quantitate Rgg-regulated transcripts from NZ131 wild-type and speB and rgg mutant strains. The results obtained with RT-PCR correlated with the proteomic data. We conclude that Rgg regulates the transcription of several genes expressed primarily during the stationary phase of growth.
Figures



References
-
- Berge A, Björck L. Streptococcal cysteine proteinase releases biologically active fragments of streptococcal surface proteins. J Biol Chem. 1995;270:9862–9867. - PubMed
-
- Bernish B, van de Rijn I. Characterization of a two-component system in Streptococcus pyogenes which is involved in regulation of hyaluronic acid production. J Biol Chem. 1999;274:4786–4793. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

