Microsporidian invasion apparatus: identification of a novel polar tube protein and evidence for clustering of ptp1 and ptp2 genes in three Encephalitozoon species
- PMID: 11159998
- PMCID: PMC97982
- DOI: 10.1128/IAI.69.2.1016-1024.2001
Microsporidian invasion apparatus: identification of a novel polar tube protein and evidence for clustering of ptp1 and ptp2 genes in three Encephalitozoon species
Abstract
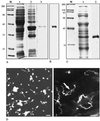
Microsporidia are unicellular eukaryotes occurring as obligate intracellular parasites which produce resistant spores. A unique motile process is represented by the sudden extrusion of the sporal polar tube for initiating entry of the parasite into a new host cell. The complete sequence of an acidic proline-rich polar tube protein (renamed PTP1) has been previously reported for Encephalitozoon cuniculi and E. hellem. Our immunological investigations provided evidence for an additional PTP in E. cuniculi, termed PTP2. The corresponding gene was sequenced and then expressed in Escherichia coli. As expected, mouse antibodies raised against the recombinant protein reacted specifically with the polar tube. The single copy ptp1 and ptp2 genes of E. cuniculi were tandemly arranged on chromosome VI. Polyadenylation of the mRNAs was demonstrated. Identification and sequencing of homologous genes in the two other human-infecting Encephalitozoon species (ptp2 in E. hellem and ptp1 and ptp2 in E. intestinalis) were facilitated by conserved gene clustering. PTP2 appears as a novel structural protein (30 kDa) with a basic lysine-rich core and an acidic tail. Unlike PTP1, this protein is devoid of large tandem repeats. The interspecies conservation of cysteine residues supports a major role of disulfide bridges in polar tube assembly. The two PTPs should serve as both molecular markers of spore differentiation and diagnostic tools.
Figures




References
-
- Biderre C, Pagès M, Méténier G, Canning E U, Vivarès C P. Evidence for the smallest nuclear genome (2, 9 Mb) in the microsporidium Encephalitozoon cuniculi. Mol Biochem Parasitol. 1995;74:229–231. - PubMed
-
- Biderre C, Canning E U, Méténier P G, Vivarès C P. Comparison of two isolates of Encephalitozoon hellem and E. intestinalis (Microspora) by pulsed field gel electrophoresis. Eur J Protistol. 1999;35:194–196.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

