Mechanisms governing expression of the v-FLIP gene of Kaposi's sarcoma-associated herpesvirus
- PMID: 11160684
- PMCID: PMC114095
- DOI: 10.1128/JVI.75.4.1857-1863.2001
Mechanisms governing expression of the v-FLIP gene of Kaposi's sarcoma-associated herpesvirus
Abstract
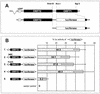
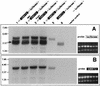
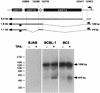
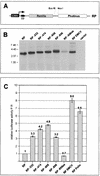
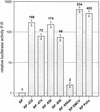
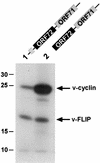
Open reading frame 71 (ORF 71) of Kaposi's sarcoma-associated herpesvirus (KSHV) encodes a death effector domain-containing protein that is homologous to cellular FLIPs (FLICE-inhibitory proteins) and is proposed to inhibit Fas-mediated apoptosis. Transcripts bearing ORF 71 (v-FLIP) sequences are present in all latently infected cells. However, mapping studies reveal these to be bi- or tricistronic mRNAs with ORF 71 located 3' to ORFs 72 (v-cyclin) and 73 (latency-associated nuclear antigen), raising the question of how efficient expression of v-FLIP is achieved. We explored this question by examining the expression of model bicistronic (v-cyclin/LUC) transcripts in which a luciferase (LUC) reporter replaced v-FLIP coding sequences. SLK spindle cells transfected with such constructs efficiently expressed luciferase from the 3' position, and this expression was independent of the expression of the 5' v-cyclin gene. Surprisingly, transcript mapping showed that in these cultures, efficient splicing occurred to remove v-cyclin sequences and generate monocistronic LUC transcripts. Similar splicing events produced monocistronic v-FLIP transcripts in KSHV-infected primary effusion lymphoma cells. However, these RNAs were of low abundance and were inducible by treatment with 12-O-tetradecanoylphorbol-13-acetate. Examination of the more abundant bicistronic latent RNAs revealed the presence of an efficient internal ribosome entry site (IRES) overlapping ORF 72 coding sequences. Thus, two potential mechanisms exist for v-FLIP expression, but the evidence suggests that IRES-mediated internal translational initiation on latent polycistronic mRNAs is the principal source of v-FLIP in latency.
Figures

References
-
- Arvanitakis L, Mesri E A, Nador R G, Said J W, Asch A S, Knowles D M, Cesarman E. Establishment and characterization of a primary effusion (body cavity-based) lymphoma cell line (BC-3) harboring Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) in the absence of Epstein-Barr virus. Blood. 1996;88:2648–2654. - PubMed
-
- Ballestas M E, Chatis P A, Kaye K M. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science. 1999;284:641–644. - PubMed
-
- Cesarman E, Chang Y, Moore P S, Said J W, Knowles D M. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. - PubMed
-
- Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266:1865–1869. - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

