Does a cdc2 kinase-like recognition motif on the core protein of hepadnaviruses regulate assembly and disintegration of capsids?
- PMID: 11160705
- PMCID: PMC115152
- DOI: 10.1128/JVI.75.4.2024-2028.2001
Does a cdc2 kinase-like recognition motif on the core protein of hepadnaviruses regulate assembly and disintegration of capsids?
Abstract
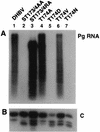
Hepadnaviruses are enveloped viruses, each with a DNA genome packaged in an icosahedral nucleocapsid, which is the site of viral DNA synthesis. In the presence of envelope proteins, DNA-containing nucleocapsids are assembled into virions and secreted, but in the absence of these proteins, nucleocapsids deliver viral DNA into the cell nucleus. Presumably, this step is identical to the delivery of viral DNA during the initiation of an infection. Unfortunately, the mechanisms triggering the disintegration of subviral core particles and delivery of viral DNA into the nucleus are not yet understood. We now report the identification of a sequence motif resembling a serine- or threonine-proline kinase recognition site in the core protein at a location that is required for the assembly of core polypeptides into capsids. Using duck hepatitis B virus, we demonstrated that mutations at this sequence motif can have profound consequences for RNA packaging, DNA replication, and core protein stability. Furthermore, we found a mutant with a conditional phenotype that depended on the cell type used for virus replication. Our results support the hypothesis predicting that this motif plays a role in assembly and disassembly of viral capsids.
Figures

References
-
- Bottcher B, Wynne S A, Crowther R A. Determination of the fold of the core protein of hepatitis B virus by electron cryomicroscopy. Nature. 1997;386:88–91. - PubMed
-
- Conway J F, Cheng N, Zlotnick A, Wingfield P T, Stahl S J, Steven A C. Visualization of a 4-helix bundle in the hepatitis B virus capsid by cryo-electron microscopy. Nature. 1997;386:91–94. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

