Ric1p and the Ypt6p GTPase function in a common pathway required for localization of trans-Golgi network membrane proteins
- PMID: 11160819
- PMCID: PMC30564
- DOI: 10.1091/mbc.12.1.13
Ric1p and the Ypt6p GTPase function in a common pathway required for localization of trans-Golgi network membrane proteins
Abstract
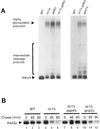
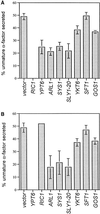
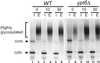
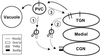
In Saccharomyces cerevisiae, clathrin is necessary for localization of trans-Golgi network (TGN) membrane proteins, a process that involves cycling of TGN proteins between the TGN and endosomes. To characterize further TGN protein localization, we applied a screen for mutations that cause severe growth defects in combination with a temperature-sensitive clathrin heavy chain. This screen yielded a mutant allele of RIC1. Cells carrying a deletion of RIC1 (ric1Delta) mislocalize TGN membrane proteins Kex2p and Vps10p to the vacuole. Delivery to the vacuole occurs in ric1Delta cells also harboring end3Delta to block endocytosis, indicative of a defect in retrieval to the TGN rather than sorting to endosomes. SYS1, originally discovered as a multicopy suppressor of defects caused by the absence of the Rab GTPase YPT6, was identified as a multicopy suppressor of ric1Delta. Further comparison of ric1Delta and ypt6Delta cells demonstrated identical phenotypes. Multicopy plasmids expressing v-SNAREs Gos1p or Ykt6p, but not other v- and t-SNAREs, partially suppressed phenotypes of ric1Delta and ypt6Delta cells. SLY1-20, a dominant activator of the cis-Golgi network t-SNARE Sed5p, also functioned as a multicopy suppressor. Because Gos1p and Ykt6p interact with Sed5p, these results raise the possibility that TGN membrane protein localization requires Ric1p- and Ypt6p-dependent retrieval to the cis-Golgi network.
Figures

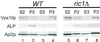


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

