KH domain: one motif, two folds
- PMID: 11160884
- PMCID: PMC30387
- DOI: 10.1093/nar/29.3.638
KH domain: one motif, two folds
Abstract
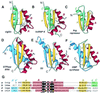
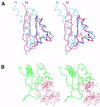
The K homology (KH) module is a widespread RNA-binding motif that has been detected by sequence similarity searches in such proteins as heterogeneous nuclear ribonucleoprotein K (hnRNP K) and ribosomal protein S3. Analysis of spatial structures of KH domains in hnRNP K and S3 reveals that they are topologically dissimilar and thus belong to different protein folds. Thus KH motif proteins provide a rare example of protein domains that share significant sequence similarity in the motif regions but possess globally distinct structures. The two distinct topologies might have arisen from an ancestral KH motif protein by N- and C-terminal extensions, or one of the existing topologies may have evolved from the other by extension, displacement and deletion. C-terminal extension (deletion) requires ss-sheet rearrangement through the insertion (removal) of a ss-strand in a manner similar to that observed in serine protease inhibitors serpins. Current analysis offers a new look on how proteins can change fold in the course of evolution.
Figures
References
-
- Hubbard T.J. and Blundell,T.L. (1987) Comparison of solvent-inaccessible cores of homologous proteins: definitions useful for protein modelling. Protein Eng., 1, 159–171. - PubMed
-
- Grishin N.V. (1997) Estimation of evolutionary distances from protein spatial structures. J. Mol. Evol., 45, 359–369. - PubMed
-
- Doolittle R.F. (1981) Similar amino acid sequences: chance or common ancestry? Science, 214, 149–159. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

