Antisense-mediated decrease in DNA ligase III expression results in reduced mitochondrial DNA integrity
- PMID: 11160888
- PMCID: PMC30390
- DOI: 10.1093/nar/29.3.668
Antisense-mediated decrease in DNA ligase III expression results in reduced mitochondrial DNA integrity
Abstract
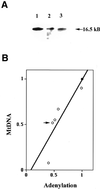
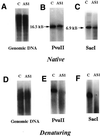
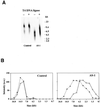
The human DNA ligase III gene encodes both nuclear and mitochondrial proteins. Abundant evidence supports the conclusion that the nuclear DNA ligase III protein plays an essential role in both base excision repair and homologous recombination. However, the role of DNA ligase III protein in mitochondrial genome dynamics has been obscure. Human tumor-derived HT1080 cells were transfected with an antisense DNA ligase III expression vector and clones with diminished levels of DNA ligase III activity identified. Mitochondrial protein extracts prepared from these clones had decreased levels of DNA ligase III relative to extracts from cells transfected with a control vector. Analysis of these clones revealed that the DNA ligase III antisense mRNA-expressing cells had reduced mtDNA content compared to control cells. In addition, the residual mtDNA present in these cells had numerous single-strand nicks that were not detected in mtDNA from control cells. Cells expressing antisense ligase III also had diminished capacity to restore their mtDNA to pre-irradiation levels following exposure to gamma-irradiation. An antisense-mediated reduction in cellular DNA ligase IV had no effect on the copy number or integrity of mtDNA. This observation, coupled with other evidence, suggests that DNA ligase IV is not present in the mitochondria and does not play a role in maintaining mtDNA integrity. We conclude that DNA ligase III is essential for the proper maintenance of mtDNA in cultured mammalian somatic cells.
Figures




References
-
- Applegren N., Hickey,R.J., Kleinschmidt,A.M., Zhou,Q.Q., Cell,J., Wills,P., Swaby,R., Wei,Y.T., Quan,J.Y., Lee,M.Y.W.T. and Malkas,L.H. (1995) Further characterization of the human cell multiprotein DNA replication complex. J. Cell. Biochem., 59, 91–107. - PubMed
-
- Waga S., Bauer,G. and Stillman,B. (1994) Reconstitution of complete SV40 replication with purified replication factors. J. Biol. Chem., 69, 10923–10934. - PubMed
-
- Aboussekhra A., Biggerstaff,M., Shivji,M.K.K., Vilpo,J.A., Moncollin,V., Podust,V.N., Protic,M., Hubscher,U., Egly,J.M. and Wood,R.D. (1995) Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell, 80, 859–868. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

