Diversity arrays: a solid state technology for sequence information independent genotyping
- PMID: 11160945
- PMCID: PMC29632
- DOI: 10.1093/nar/29.4.e25
Diversity arrays: a solid state technology for sequence information independent genotyping
Abstract
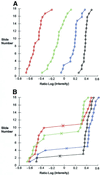
Here we present the successful application of the microarray technology platform to the analysis of DNA polymorphisms. Using the rice genome as a model, we demonstrate the potential of a high-throughput genome analysis method called Diversity Array Technology, DArT'. In the format presented here the technology is assaying for the presence (or amount) of a specific DNA fragment in a representation derived from the total genomic DNA of an organism or a population of organisms. Two different approaches are presented: the first involves contrasting two representations on a single array while the second involves contrasting a representation with a reference DNA fragment common to all elements of the array. The Diversity Panels created using this method allow genetic fingerprinting of any organism or group of organisms belonging to the gene pool from which the panel was developed. Diversity Arrays enable rapid and economical application of a highly parallel, solid-state genotyping technology to any genome or complex genomic mixtures.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

