Dynamic regulation of the tryptophan operon: a modeling study and comparison with experimental data
- PMID: 11171956
- PMCID: PMC29262
- DOI: 10.1073/pnas.98.4.1364
Dynamic regulation of the tryptophan operon: a modeling study and comparison with experimental data
Abstract
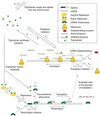
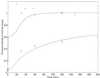
A mathematical model for regulation of the tryptophan operon is presented. This model takes into account repression, feedback enzyme inhibition, and transcriptional attenuation. Special attention is given to model parameter estimation based on experimental data. The model's system of delay differential equations is numerically solved, and the results are compared with experimental data on the temporal evolution of enzyme activity in cultures of Escherichia coli after a nutritional shift (minimal + tryptophan medium to minimal medium). Good agreement is obtained between the numeric simulations and the experimental results for wild-type E. coli, as well as for two different mutant strains.
Figures


References
-
- Jacob F, Perrin D, Sanchez C, Monod J. C R Seances Acad Sci. 1960;250:1727–1729. - PubMed
-
- Beckwith J. In: Escherichia coli and Salmonella thyphymurium: Cellular and Molecular Biology. Neidhart F C, Curtiss R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Vol. 2. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 1553–1569.
-
- Goodwin B. Adv Enzyme Regul. 1965;3:425–438. - PubMed
-
- Bliss R D, Painter R P, Marr A G. J Theor Biol. 1982;97:177–193. - PubMed
-
- Sinha S. Biotechnol Bioeng. 1988;31:117–124. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

