Interaction of the herbicide glyphosate with its target enzyme 5-enolpyruvylshikimate 3-phosphate synthase in atomic detail
- PMID: 11171958
- PMCID: PMC29264
- DOI: 10.1073/pnas.98.4.1376
Interaction of the herbicide glyphosate with its target enzyme 5-enolpyruvylshikimate 3-phosphate synthase in atomic detail
Abstract
Biosynthesis of aromatic amino acids in plants, many bacteria, and microbes relies on the enzyme 5-enolpyruvylshikimate 3-phosphate (EPSP) synthase, a prime target for drugs and herbicides. We have identified the interaction of EPSP synthase with one of its two substrates (shikimate 3-phosphate) and with the widely used herbicide glyphosate by x-ray crystallography. The two-domain enzyme closes on ligand binding, thereby forming the active site in the interdomain cleft. Glyphosate appears to occupy the binding site of the second substrate of EPSP synthase (phosphoenol pyruvate), mimicking an intermediate state of the ternary enzyme.substrates complex. The elucidation of the active site of EPSP synthase and especially of the binding pattern of glyphosate provides a valuable roadmap for engineering new herbicides and herbicide-resistant crops, as well as new antibiotic and antiparasitic drugs.
Figures
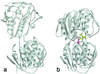
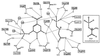
Comment in
-
Closing down on glyphosate inhibition--with a new structure for drug discovery.Proc Natl Acad Sci U S A. 2001 Mar 13;98(6):2944-6. doi: 10.1073/pnas.061025898. Proc Natl Acad Sci U S A. 2001. PMID: 11248008 Free PMC article. Review. No abstract available.
References
-
- Bentley R. Crit Rev Biochem Mol Biol. 1990;25:307–383. - PubMed
-
- Haslam E. Shicimic Acid: Metabolism and Metabolites. Chichester, U.K.: Wiley; 1993.
-
- Kishore G M, Shah D M. Annu Rev Biochem. 1988;57:627–663. - PubMed
-
- Roberts F, Roberts C W, Johnson J J, Kyle D E, Krell T, Coggins J R, Coombs G H, Milhous W K, Tzipori S, Ferguson D J P, et al. Nature (London) 1998;393:801–805. - PubMed
-
- Steinrücken H C, Amrhein N. Biochem Biophys Res Commun. 1980;94:1207–1212. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

