Involvement of a cytosine side chain in proton transfer in the rate-determining step of ribozyme self-cleavage
- PMID: 11171978
- PMCID: PMC29284
- DOI: 10.1073/pnas.98.4.1489
Involvement of a cytosine side chain in proton transfer in the rate-determining step of ribozyme self-cleavage
Abstract
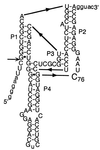
Ribozymes of hepatitis delta virus have been proposed to use an active-site cytosine as an acid-base catalyst in the self-cleavage reaction. In this study, we have examined the role of cytosine in more detail with the antigenomic ribozyme. Evidence that proton transfer in the rate-determining step involved cytosine 76 (C76) was obtained from examining cleavage activity of the wild-type and imidazole buffer-rescued C76-deleted (C76 Delta) ribozymes in D(2)O and H(2)O. In both reactions, a similar kinetic isotope effect and shift in the apparent pKa indicate that the buffer is functionally substituting for the side chain in proton transfer. Proton inventory of the wild-type reaction supported a mechanism of a single proton transfer at the transition state. This proton transfer step was further characterized by exogenous base rescue of a C76 Delta mutant with cytosine and imidazole analogues. For the imidazole analogues that rescued activity, the apparent pKa of the rescue reaction, measured under k(cat)/K(M) conditions, correlated with the pKa of the base. From these data a Brønsted coefficient (beta) of 0.51 was determined for the base-rescued reaction of C76 Delta. This value is consistent with that expected for proton transfer in the transition state. Together, these data provide strong support for a mechanism where an RNA side chain participates directly in general acid or general base catalysis of the wild-type ribozyme to facilitate RNA cleavage.
Figures



References
-
- Jencks W P. Adv Enzymol Relat Areas Mol Biol. 1975;43:219–410. - PubMed
-
- Toney M D, Kirsch J F. Science. 1989;243:1485–1488. - PubMed
-
- Huang S, Tu S C. Biochemistry. 1997;36:14609–14615. - PubMed
-
- Newmyer S L, de Montellano P R O. J Biol Chem. 1996;271:14891–14896. - PubMed
-
- Brown R S, Dewan J C, Klug A. Biochemistry. 1985;24:4785–4801. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

