Identification and modulation of a naturally processed T cell epitope from the diabetes-associated autoantigen human glutamic acid decarboxylase 65 (hGAD65)
- PMID: 11172025
- PMCID: PMC29331
- DOI: 10.1073/pnas.98.4.1763
Identification and modulation of a naturally processed T cell epitope from the diabetes-associated autoantigen human glutamic acid decarboxylase 65 (hGAD65)
Abstract
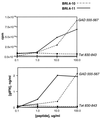
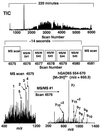
T cell recognition of autoantigens is critical to progressive immune-mediated destruction of islet cells, which leads to autoimmune diabetes. We identified a naturally presented autoantigen from the human islet antigen glutamic acid decarboxylase, 65-kDa isoform (GAD65), by using a combination of chromatography and mass spectrometry of peptides bound by the type I diabetes (insulin-dependent diabetes mellitus, IDDM)-associated HLA-DR4 molecule. Peptides encompassing this epitope-stimulated GAD65-specific T cells from diabetic patients and a DR4-positive individual at high risk for developing IDDM. T cell responses were antagonized by altered peptide ligands containing single amino acid modifications. This direct identification and manipulation of GAD65 epitope recognition provides an approach toward dissection of the complex CD4(+) T cell response in IDDM.
Figures



References
-
- Gianani R, Eisenbarth G S. In: Type I Diabetes. Molecular, Cellular, and Clinical Immunology. Eisenbarth G S, Lafferty K J, editors. New York: Oxford Univ. Press; 1996. pp. 209–229.
-
- Nepom G T. Curr Opin Immunol. 1995;7:825–830. - PubMed
-
- Mehta V, Palmer J P. In: Prediction, Prevention and Genetic Counseling in IDDM. Palmer J P, editor. Chichester, PA: Wiley; 1996. pp. 3–16.
-
- Lernmark A. J Intern Med. 1996;240:259–277. - PubMed
-
- Yoon J W, Yoon C S, Lim H W, Huang Q Q, Kang Y, Pyun K H, Hirasawa K, Sherwin R S, Jun H S. Science. 1999;284:1183–1187. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

