An overview of the potassium channel family
- PMID: 11178249
- PMCID: PMC138870
- DOI: 10.1186/gb-2000-1-4-reviews0004
An overview of the potassium channel family
Abstract
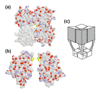
Potassium channels, tetrameric integral membrane proteins that form aqueous pores through which K+ can flow, are found in virtually all organisms; the genomes of humans, Drosophila, and Caenorhabditis elegans contain 30-100 K+ channel genes each. The structure of a bacterial K+ channel, sequence comparisons with other channels and electrophysiological measurements have enabled conclusions about the mechanism of gating and ion flow to be drawn for many other channels.
Figures


References
-
- Hille B. Ionic Channels of Excitable Membranes. . Sunderland, Massachusetts: Sinauer Associates, 1992.
-
- Littleton JT, Ganetzky B. Ion channels and synaptic organization: analysis of the Drosophila genome. . Neuron. 2000;26:35–43. - PubMed
-
- Papazian DM, Schwarz TL, Tempel BL, Jan YN, Jan LY. Cloning of genomic and complementary DNA from Shaker, a putative potassium channel gene from Drosophila. Science. 1987;237:749–753. - PubMed
-
- Ho K, Nichols CG, Lederer WJ, Lytton J, Vassilev PM, Kanazirska MV, Hebert SC. Cloning and expression of an inwardly rectifying ATP-regulated potassium channel. Nature. 1993;362:31–38. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

