Towards understanding the first genome sequence of a crenarchaeon by genome annotation using clusters of orthologous groups of proteins (COGs)
- PMID: 11178258
- PMCID: PMC15027
- DOI: 10.1186/gb-2000-1-5-research0009
Towards understanding the first genome sequence of a crenarchaeon by genome annotation using clusters of orthologous groups of proteins (COGs)
Abstract
Background: Standard archival sequence databases have not been designed as tools for genome annotation and are far from being optimal for this purpose. We used the database of Clusters of Orthologous Groups of proteins (COGs) to reannotate the genomes of two archaea, Aeropyrum pernix, the first member of the Crenarchaea to be sequenced, and Pyrococcus abyssi.
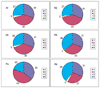
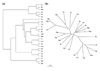
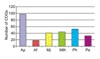
Results: A. pernix and P. abyssi proteins were assigned to COGs using the COGNITOR program; the results were verified on a case-by-case basis and augmented by additional database searches using the PSI-BLAST and TBLASTN programs. Functions were predicted for over 300 proteins from A. pernix, which could not be assigned a function using conventional methods with a conservative sequence similarity threshold, an approximately 50% increase compared to the original annotation. A. pernix shares most of the conserved core of proteins that were previously identified in the Euryarchaeota. Cluster analysis or distance matrix tree construction based on the co-occurrence of genomes in COGs showed that A. pernix forms a distinct group within the archaea, although grouping with the two species of Pyrococci, indicative of similar repertoires of conserved genes, was observed. No indication of a specific relationship between Crenarchaeota and eukaryotes was obtained in these analyses. Several proteins that are conserved in Euryarchaeota and most bacteria are unexpectedly missing in A. pernix, including the entire set of de novo purine biosynthesis enzymes, the GTPase FtsZ (a key component of the bacterial and euryarchaeal cell-division machinery), and the tRNA-specific pseudouridine synthase, previously considered universal. A. pernix is represented in 48 COGs that do not contain any euryarchaeal members. Many of these proteins are TCA cycle and electron transport chain enzymes, reflecting the aerobic lifestyle of A. pernix.
Conclusions: Special-purpose databases organized on the basis of phylogenetic analysis and carefully curated with respect to known and predicted protein functions provide for a significant improvement in genome annotation. A differential genome display approach helps in a systematic investigation of common and distinct features of gene repertoires and in some cases reveals unexpected connections that may be indicative of functional similarities between phylogenetically distant organisms and of lateral gene exchange.
Figures

Similar articles
-
Clusters of orthologous genes for 41 archaeal genomes and implications for evolutionary genomics of archaea.Biol Direct. 2007 Nov 27;2:33. doi: 10.1186/1745-6150-2-33. Biol Direct. 2007. PMID: 18042280 Free PMC article.
-
Phylogenomic analysis of proteins that are distinctive of Archaea and its main subgroups and the origin of methanogenesis.BMC Genomics. 2007 Mar 29;8:86. doi: 10.1186/1471-2164-8-86. BMC Genomics. 2007. PMID: 17394648 Free PMC article.
-
Updated clusters of orthologous genes for Archaea: a complex ancestor of the Archaea and the byways of horizontal gene transfer.Biol Direct. 2012 Dec 14;7:46. doi: 10.1186/1745-6150-7-46. Biol Direct. 2012. PMID: 23241446 Free PMC article.
-
Archaeal genomics.Curr Opin Microbiol. 1999 Oct;2(5):542-7. doi: 10.1016/s1369-5274(99)00014-4. Curr Opin Microbiol. 1999. PMID: 10508726 Review.
-
Status of genome projects for nonpathogenic bacteria and archaea.Nat Biotechnol. 2000 Oct;18(10):1049-54. doi: 10.1038/80235. Nat Biotechnol. 2000. PMID: 11017041 Review.
Cited by
-
Complete Mitogenomes of Three Carangidae (Perciformes) Fishes: Genome Description and Phylogenetic Considerations.Int J Mol Sci. 2020 Jun 30;21(13):4685. doi: 10.3390/ijms21134685. Int J Mol Sci. 2020. PMID: 32630142 Free PMC article.
-
The COG database: new developments in phylogenetic classification of proteins from complete genomes.Nucleic Acids Res. 2001 Jan 1;29(1):22-8. doi: 10.1093/nar/29.1.22. Nucleic Acids Res. 2001. PMID: 11125040 Free PMC article.
-
Revealing gene transcription and translation initiation patterns in archaea, using an interactive clustering model.Extremophiles. 2004 Aug;8(4):291-9. doi: 10.1007/s00792-004-0388-1. Epub 2004 May 19. Extremophiles. 2004. PMID: 15150699
-
Lineage-specific gene expansions in bacterial and archaeal genomes.Genome Res. 2001 Apr;11(4):555-65. doi: 10.1101/gr.gr-1660r. Genome Res. 2001. PMID: 11282971 Free PMC article.
-
Flower transcriptome dynamics during nectary development in pepper (Capsicum annuum L.).Genet Mol Biol. 2020 May 29;43(2):e20180267. doi: 10.1590/1678-4685-GMB-2018-0267. eCollection 2020. Genet Mol Biol. 2020. PMID: 32478788 Free PMC article.
References
-
- Boguski MS. Biosequence exegesis. Science. 1999;286:453–455. - PubMed
-
- Bork P, Dandekar T, Diaz-Lazcoz Y, Eisenhaber F, Huynen M, Yuan Y. Predicting function: from genes to genomes and back. J Mol Biol. 1998;283:707–725. - PubMed
-
- Galperin MY, Koonin EV. Who's your neighbor? New computational approaches for functional genomics. Nat Biotechnol. 2000;18:609–613. - PubMed
-
- Andrade MA, Brown NP, Leroy C, Hoersch S, de Daruvar A, Reich C, Franchini A, Tamames J, Valencia A, Ouzounis C, Sander C. Automated genome sequence analysis and annotation. Bioinformatics. 1999;15:391–412. - PubMed
-
- Gaasterland T, Sensen CW. MAGPIE: automated genome interpretation. Trends Genet. 1996;12:76–78. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

