Evidence for symmetric chromosomal inversions around the replication origin in bacteria
- PMID: 11178265
- PMCID: PMC16139
- DOI: 10.1186/gb-2000-1-6-research0011
Evidence for symmetric chromosomal inversions around the replication origin in bacteria
Abstract
Background: Whole-genome comparisons can provide great insight into many aspects of biology. Until recently, however, comparisons were mainly possible only between distantly related species. Complete genome sequences are now becoming available from multiple sets of closely related strains or species.
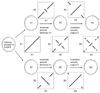
Results: By comparing the recently completed genome sequences of Vibrio cholerae, Streptococcus pneumoniae and Mycobacterium tuberculosis to those of closely related species - Escherichia coli, Streptococcus pyogenes and Mycobacterium leprae, respectively - we have identified an unusual and previously unobserved feature of bacterial genome structure. Scatterplots of the conserved sequences (both DNA and protein) between each pair of species produce a distinct X-shaped pattern, which we call an X-alignment. The key feature of these alignments is that they have symmetry around the replication origin and terminus; that is, the distance of a particular conserved feature (DNA or protein) from the replication origin (or terminus) is conserved between closely related pairs of species. Statistically significant X-alignments are also found within some genomes, indicating that there is symmetry about the replication origin for paralogous features as well.
Conclusions: The most likely mechanism of generation of X-alignments involves large chromosomal inversions that reverse the genomic sequence symmetrically around the origin of replication. The finding of these X-alignments between many pairs of species suggests that chromosomal inversions around the origin are a common feature of bacterial genome evolution.
Figures



Comment in
-
Flip-flop around the origin and terminus of replication in prokaryotic genomes.Genome Biol. 2001;2(12):INTERACTIONS1004. doi: 10.1186/gb-2001-2-12-interactions1004. Epub 2001 Nov 15. Genome Biol. 2001. PMID: 11790247 Free PMC article.
Similar articles
-
Replication-associated inversions are the dominant form of bacterial chromosome structural variation.Life Sci Alliance. 2022 Oct 19;6(1):e202201434. doi: 10.26508/lsa.202201434. Print 2023 Jan. Life Sci Alliance. 2022. PMID: 36261227 Free PMC article.
-
Insertion sequence elements and unique symmetrical genomic regions mediate chromosomal inversions in Streptococcus pyogenes.Nucleic Acids Res. 2024 Nov 27;52(21):13128-13137. doi: 10.1093/nar/gkae948. Nucleic Acids Res. 2024. PMID: 39460626 Free PMC article.
-
In silico prediction of the origin of replication among bacteria: a case study of Bacteroides thetaiotaomicron.OMICS. 2008 Sep;12(3):201-10. doi: 10.1089/omi.2008.0004. OMICS. 2008. PMID: 18582175
-
End of the beginning: elongation and termination features of alternative modes of chromosomal replication initiation in bacteria.PLoS Genet. 2015 Jan 8;11(1):e1004909. doi: 10.1371/journal.pgen.1004909. eCollection 2015 Jan. PLoS Genet. 2015. PMID: 25569209 Free PMC article. Review.
-
Management of multipartite genomes: the Vibrio cholerae model.Curr Opin Microbiol. 2014 Dec;22:120-6. doi: 10.1016/j.mib.2014.10.003. Curr Opin Microbiol. 2014. PMID: 25460805 Review.
Cited by
-
Gut symbionts from distinct hosts exhibit genotoxic activity via divergent colibactin biosynthesis pathways.Appl Environ Microbiol. 2015 Feb;81(4):1502-12. doi: 10.1128/AEM.03283-14. Appl Environ Microbiol. 2015. PMID: 25527542 Free PMC article.
-
Azotobacter Genomes: The Genome of Azotobacter chroococcum NCIMB 8003 (ATCC 4412).PLoS One. 2015 Jun 10;10(6):e0127997. doi: 10.1371/journal.pone.0127997. eCollection 2015. PLoS One. 2015. PMID: 26061173 Free PMC article.
-
A blueprint of ectoine metabolism from the genome of the industrial producer Halomonas elongata DSM 2581 T.Environ Microbiol. 2011 Aug;13(8):1973-94. doi: 10.1111/j.1462-2920.2010.02336.x. Epub 2010 Sep 16. Environ Microbiol. 2011. PMID: 20849449 Free PMC article.
-
The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome.Proc Natl Acad Sci U S A. 2008 Dec 2;105(48):18964-9. doi: 10.1073/pnas.0809584105. Epub 2008 Nov 24. Proc Natl Acad Sci U S A. 2008. PMID: 19033196 Free PMC article.
-
DNA motifs that sculpt the bacterial chromosome.Nat Rev Microbiol. 2011 Jan;9(1):15-26. doi: 10.1038/nrmicro2477. Nat Rev Microbiol. 2011. PMID: 21164534 Review.
References
-
- Seoighe C, Wolfe KH. Updated map of duplicated regions in the yeast genome. Gene. 1999;238:253–261. - PubMed
-
- Lin X, Kaul S, Rounsley S, Shea TP, Benito MI, Town CD, Fujii CY, Mason T, Bowman CL, Barnstead M, et al. Sequence and analysis of chromosome 2 of the plant Arabidopsis thaliana. Nature. 1999;402:761–768. - PubMed
-
- Mayer K, Schuller C, Wambutt R, Murphy G, Volckaert G, Pohl T, Dusterhoft A, Stiekema W, Entian KD, Terryn N, et al. Sequence and analysis of chromosome 4 of the plant Arabidopsis thaliana. Nature. 1999;402:769–777. - PubMed
-
- Blattner FR, Plunkett GI, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

