Genome sequences and great expectations
- PMID: 11178275
- PMCID: PMC150431
- DOI: 10.1186/gb-2000-2-1-interactions0001
Genome sequences and great expectations
Abstract
To assess how automatic function assignment will contribute to genome annotation in the next five years, we have performed an analysis of 31 available genome sequences. An emerging pattern is that function can be predicted for almost two-thirds of the 73,500 genes that were analyzed. Despite progress in computational biology, there will always be a great need for large-scale experimental determination of protein function.
Figures
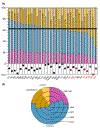
References
-
- Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR, Bult CJ, Tomb JF, Dougherty BA, Merrick JM, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. - PubMed
-
- Casari G, Andrade MA, Bork P, Boyle J, Daruvar A, Ouzounis C, Schneider R, Tamames J, Valencia A, Sander C. Challenging times for bioinformatics. Nature. 1995;376:647–648. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

