Supervised harvesting of expression trees
- PMID: 11178280
- PMCID: PMC17599
- DOI: 10.1186/gb-2001-2-1-research0003
Supervised harvesting of expression trees
Abstract
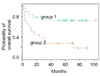
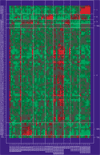
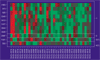
Background: We propose a new method for supervised learning from gene expression data. We call it 'tree harvesting'. This technique starts with a hierarchical clustering of genes, then models the outcome variable as a sum of the average expression profiles of chosen clusters and their products. It can be applied to many different kinds of outcome measures such as censored survival times, or a response falling in two or more classes (for example, cancer classes). The method can discover genes that have strong effects on their own, and genes that interact with other genes.
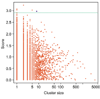
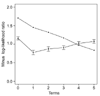
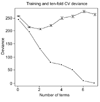
Results: We illustrate the method on data from a lymphoma study, and on a dataset containing samples from eight different cancers. It identified some potentially interesting gene clusters. In simulation studies we found that the procedure may require a large number of experimental samples to successfully discover interactions.
Conclusions: Tree harvesting is a potentially useful tool for exploration of gene expression data and identification of interesting clusters of genes worthy of further investigation.
Figures


References
-
- Friedman J. Multivariate adaptive regression splines. Annl Stat. 1991;19:1–141.
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

