Compound heterozygosity for a recurrent 16.5-kb Alu-mediated deletion mutation and single-base-pair substitutions in the ABCC6 gene results in pseudoxanthoma elasticum
- PMID: 11179012
- PMCID: PMC1274477
- DOI: 10.1086/318807
Compound heterozygosity for a recurrent 16.5-kb Alu-mediated deletion mutation and single-base-pair substitutions in the ABCC6 gene results in pseudoxanthoma elasticum
Abstract
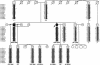
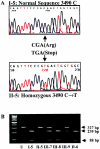
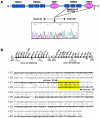
Pseudoxanthoma elasticum (PXE) is a systemic heritable disorder affecting the elastic structures in the skin, eyes, and cardiovascular system, with considerable morbidity and mortality. Recently, mutations in the ABCC6 gene (also referred to as "MRP6" or "eMOAT") encoding multidrug-resistance protein 6 (MRP6), a putative transmembrane ABC transporter protein of unknown function, have been disclosed. Most of the genetic lesions delineated thus far consist of single-base-pair substitutions resulting in nonsense, missense, or splice-site mutations. In this study, we examined four multiplex families with PXE inherited in an autosomal recessive pattern. In each family, the proband was a compound heterozygote for a single-base-pair-substitution mutation and a novel, approximately 16.5-kb deletion mutation spanning the site of the single-base-pair substitution in trans. The deletion mutation was shown to extend from intron 22 to intron 29, resulting in out-of-frame deletion of 1,213 nucleotides from the corresponding mRNA and causing elimination of 505 amino acids from the MRP6 polypeptide. The deletion breakpoints were precisely the same in all four families, which were of different ethnic backgrounds, and haplotype analysis by 13 microsatellite markers suggested that the deletion had occurred independently. Deletion breakpoints within introns 22 and 29 were embedded within AluSx repeat sequences, specifically in a 16-bp segment of DNA, suggesting Alu-mediated homologous recombination as a mechanism.
Figures

References
Electronic-Database Information
-
- GenBank Overview, http://www.ncbi.nlm.nih.gov/Genbank/GenbankOverview.html (for BAC clone A-962B4 [accession number U91318], published cDNA sequence [accession number AF076622], and nucleotides 2997–4208 of exons 23–29 [accession number AF168791])
-
- Genome Database, The, http://www.gdb.org (for microsatellite markers)
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for PXE [MIM 177850 and MIM 264800] and for Dubin-Johnson syndrome [MIM 237500])
-
- RepeatMasker Documentation, http://ftp.genome.washington.edu/RM/RepeatMasker.html
References
-
- Altman LK, Fialkow PJ, Parker F, Sagebiel RW (1974) Pseudoxanthoma elasticum: an underdiagnosed genetically heterogeneous disorder with protean manifestations. Arch Intern Med 134:1048–1054 - PubMed
-
- Bacchelli B, Quaglino D, Gheduzzi D, Taparelli F, Boraldi F, Trolli B, Le Saux O, Boyd CD, Pasquali-Ronchetti I (1999) Identification of heterozygote carriers in families with a recessive form of pseudoxanthoma elasticum (PXE). Mod Pathol 12:1112–1123 - PubMed
-
- Bergen AAB, Plomp AS, Schuurman EJ, Terry S, Breuning M, Dauwerse H, Swart J, Kool M, van Doest S, Baas F, ten Brink JB, de Jong PT (2000) Mutations in ABCC6 cause pseudoxanthoma elasticum. Nat Genet 25:228–231 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

