Crystal structure of dihydropyrimidine dehydrogenase, a major determinant of the pharmacokinetics of the anti-cancer drug 5-fluorouracil
- PMID: 11179210
- PMCID: PMC145429
- DOI: 10.1093/emboj/20.4.650
Crystal structure of dihydropyrimidine dehydrogenase, a major determinant of the pharmacokinetics of the anti-cancer drug 5-fluorouracil
Abstract
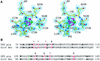
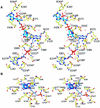
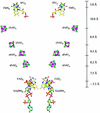
Dihydropyrimidine dehydrogenase catalyzes the first step in pyrimidine degradation: the NADPH-dependent reduction of uracil and thymine to the corresponding 5,6-dihydropyrimidines. Its controlled inhibition has become an adjunct target for cancer therapy, since the enzyme is also responsible for the rapid breakdown of the chemotherapeutic drug 5-fluorouracil. The crystal structure of the homodimeric pig liver enzyme (2x 111 kDa) determined at 1.9 A resolution reveals a highly modular subunit organization, consisting of five domains with different folds. Dihydropyrimidine dehydrogenase contains two FAD, two FMN and eight [4Fe-4S] clusters, arranged in two electron transfer chains that pass the dimer interface twice. Two of the Fe-S clusters show a hitherto unobserved coordination involving a glutamine residue. The ternary complex of an inactive mutant of the enzyme with bound NADPH and 5-fluorouracil reveals the architecture of the substrate-binding sites and residues responsible for recognition and binding of the drug.
Figures
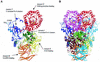



References
-
- Ahmed F.Y. et al. (1999) Eniluracil treatment completely inactivates dihydropyrimidine dehydrogenase in colorectal tumors. J. Clin. Oncol., 17, 2439–2445. - PubMed
-
- Albin N., Johnson,M.R. and Diasio,R.B. (1996) cDNA cloning of bovine liver dihydropyrimidine dehydrogenase. DNA Seq., 6, 243–250. - PubMed
-
- Brito R.A., Medgyesy,D., Zukowski,T.H., Royce,M.E., Ravandi-Kashani,F., Hoff,P.M. and Pazdur,R. (1999) Fluoropyrimidines: a critical evaluation. Oncology, 57 (suppl. 1), 2–8. - PubMed
-
- Brünger A.T. et al. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. - PubMed
-
- Bruschi M. and Guerlesquin,F. (1988) Structure, function and evolution of bacterial ferredoxins. FEMS Microbiol. Rev., 54, 155–176. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

