Molecular characterization of a novel Staphylococcus aureus serine protease operon
- PMID: 11179322
- PMCID: PMC98051
- DOI: 10.1128/IAI.69.3.1521-1527.2001
Molecular characterization of a novel Staphylococcus aureus serine protease operon
Abstract
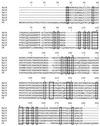
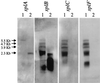
The present study identified and characterized a unique operon (spl) encoding six serine protease-like proteins. In addition, native Spl proteins were isolated and characterized. Typical of most exoproteins, the spl gene products contain putative 35- or 36-amino-acid signal peptides. The Spl proteins share 44 to 95% amino acid sequence identity with each other and 33 to 36% sequence identity with V8 protease. They also contain amino acids found in catalytic triads of enzymes in the trypsin-like serine protease family, and SplB and SplC were shown to degrade casein. The spl operon is transcribed on a 5.5-kb transcript, but several nonrandom degradation products of this transcript were also identified. Similar to other S. aureus exoprotein genes, the spl operon is maximally expressed during the transition into stationary phase and is positively controlled by the Agr virulence factor regulator. The Sar regulatory system did not affect spl operon expression. PCR analysis revealed the presence of the spl operon in 64% of the S. aureus isolates tested, although one spl operon-negative isolate was shown to contain at least two of the spl genes. Finally, intraperitoneal injection of an spl operon deletion mutant revealed no major differences in virulence compared to the parental strain.
Figures





References
-
- Arvidson A. Extracellular proteases. In: Fischetti V A, Novick R P, Ferretti J J, Portnoy D A, Rood J I, editors. Gram-positive pathogens. Washington, D.C.: American Society for Microbiology; 2000. pp. 379–385.
-
- Arvidson S, Holme T, Lindholm B. Studies on extracellular proteolytic enzymes from Staphylococcus aureus. I. Purification and characterization of one neutral and one alkaline protease. Biochim Biophys Acta. 1973;302:135–148. - PubMed
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1991.
-
- Bailey C J, Lockhart B P, Redpath M B, Smith T P. The epidermolytic (exfoliative) toxins of Staphylococcus aureus. Med Microbiol Immunol. 1995;184:53–61. - PubMed
-
- Bayles K W, Brunskill E W, Iandolo J J, Hruska L L, Huang S, Pattee P A, Smiley B K, Yasbin R E. A genetic and molecular characterization of the recA gene from Staphylococcus aureus. Gene. 1994;147:13–20. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

