Remodeling of yeast genome expression in response to environmental changes
- PMID: 11179418
- PMCID: PMC30946
- DOI: 10.1091/mbc.12.2.323
Remodeling of yeast genome expression in response to environmental changes
Abstract
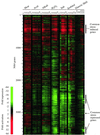
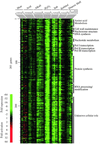
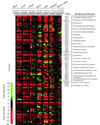
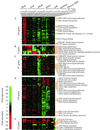
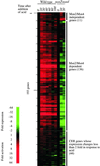
We used genome-wide expression analysis to explore how gene expression in Saccharomyces cerevisiae is remodeled in response to various changes in extracellular environment, including changes in temperature, oxidation, nutrients, pH, and osmolarity. The results demonstrate that more than half of the genome is involved in various responses to environmental change and identify the global set of genes induced and repressed by each condition. These data implicate a substantial number of previously uncharacterized genes in these responses and reveal a signature common to environmental responses that involves approximately 10% of yeast genes. The results of expression analysis with MSN2/MSN4 mutants support the model that the Msn2/Msn4 activators induce the common response to environmental change. These results provide a global description of the transcriptional response to environmental change and extend our understanding of the role of activators in effecting this response.
Figures

References
-
- Beck T, Hall MN. The TOR signaling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature. 1999;402:689–692. - PubMed
-
- Bihler H, Gaber RF, Slayman CL, Bertl A. The presumed potassium carrier Trk2p in Saccharomyces cerevisiae determines an H+-dependent, K+- independent current. FEBS Lett. 1999;447:115–120. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

