Transcript analysis of multiple copies of amo (encoding ammonia monooxygenase) and hao (encoding hydroxylamine oxidoreductase) in Nitrosomonas europaea
- PMID: 11208810
- PMCID: PMC94979
- DOI: 10.1128/JB.183.3.1096-1100.2001
Transcript analysis of multiple copies of amo (encoding ammonia monooxygenase) and hao (encoding hydroxylamine oxidoreductase) in Nitrosomonas europaea
Abstract
The genes encoding ammonia monooxygenase (amoCAB), hydroxylamine oxidoreductase (hao), and the c-type cytochrome c-554 (hcy) are present in multiple copies in the genome of Nitrosomonas europaea. The upstream regions of the two copies of amoC, the three copies of hao, and one copy of hcy were cloned and sequenced. Primer extension reactions were done to identify transcription start sites for these genes, as well as for amoA. Putative sigma(70) promoter sequences were found associated with all but one of the mapped transcription start sites. Primer extensions were done with amoC primers using RNA harvested from cells incubated with and without ammonium. The experiments suggested that N. europaea cells may be able to use different promoters in the presence and absence of ammonium.
Figures
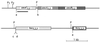

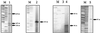

References
-
- Coburn G, Mackie G. Degradation of mRNA in Escherichia coli: an old problem with some new twists. In: Moldave K, editor. Progress in nucleic acid research and molecular biology. Vol. 62. San Diego, Calif: Academic Press; 1999. pp. 55–108. - PubMed
-
- Grafe S, Ellinger T, Malke H. Structural dissection and functional analysis of the complex promoter of the streptokinase gene from Streptococcus equisimilis H46A. Med Microbiol Immunol. 1996;185:11–17. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

