Analysis of the pmsCEAB gene cluster involved in biosynthesis of salicylic acid and the siderophore pseudomonine in the biocontrol strain Pseudomonas fluorescens WCS374
- PMID: 11222588
- PMCID: PMC95085
- DOI: 10.1128/JB.183.6.1909-1920.2001
Analysis of the pmsCEAB gene cluster involved in biosynthesis of salicylic acid and the siderophore pseudomonine in the biocontrol strain Pseudomonas fluorescens WCS374
Abstract
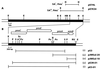
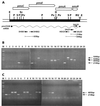
Mutants of Pseudomonas fluorescens WCS374 defective in biosynthesis of the fluorescent siderophore pseudobactin still display siderophore activity, indicating the production of a second siderophore. A recombinant cosmid clone (pMB374-07) of a WCS374 gene library harboring loci necessary for the biosynthesis of salicylic acid (SA) and this second siderophore pseudomonine was isolated. The salicylate biosynthesis region of WCS374 was localized in a 5-kb EcoRI fragment of pMB374-07. The SA and pseudomonine biosynthesis region was identified by transfer of cosmid pMB374-07 to a pseudobactin-deficient strain of P. putida. Sequence analysis of the 5-kb subclone revealed the presence of four open reading frames (ORFs). Products of two ORFs (pmsC and pmsB) showed homologies with chorismate-utilizing enzymes; a third ORF (pmsE) encoded a protein with strong similarity with enzymes involved in the biosynthesis of siderophores in other bacterial species. The region also contained a putative histidine decarboxylase gene (pmsA). A putative promoter region and two predicted iron boxes were localized upstream of pmsC. We determined by reverse transcriptase-mediated PCR that the pmsCEAB genes are cotranscribed and that expression is iron regulated. In vivo expression of SA genes was achieved in P. putida and Escherichia coli cells. In E. coli, deletions affecting the first ORF (pmsC) diminished SA production, whereas deletion of pmsB abolished it completely. The pmsB gene induced low levels of SA production in E. coli when expressed under control of the lacZ promoter. Several lines of evidence indicate that SA and pseudomonine biosynthesis are related. Moreover, we isolated a Tn5 mutant (374-05) that is simultaneously impaired in SA and pseudomonine production.
Figures

References
-
- Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Anthoni U, Christophersen C, Nielsen P H, Gram L, Petersen B O. Pseudomonine, an isoxazolidone with siderophoric activity from Pseudomonas fluorescens AH2 isolated from Lake Victorian Nile perch. J Nat Prod. 1995;58:1786–1789.
-
- Arnow L E. Colorimetric determination of the components of 3,4-dihydroxyphenylalanine-tyrosine mixtures. J Biol Chem. 1937;118:531–537.
-
- Bakker P A H M, Lamers J G, Bakker A W, Marugg J D, Weisbeek P J, Schippers B. The role of siderophores in potato tuber yield increase by Pseudomonas putida in a short rotation of potato. Neth J Plant Pathol. 1986;92:249–256.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

