Analysis of promoters recognized by PvdS, an extracytoplasmic-function sigma factor protein from Pseudomonas aeruginosa
- PMID: 11222621
- PMCID: PMC95118
- DOI: 10.1128/JB.183.6.2151-2155.2001
Analysis of promoters recognized by PvdS, an extracytoplasmic-function sigma factor protein from Pseudomonas aeruginosa
Abstract
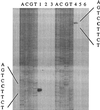
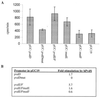
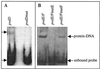
The alternative sigma factor PvdS is required by Pseudomonas aeruginosa for initiation of transcription from pyoverdine (pvd) promoters. Two divergent PvdS-dependent promoters (pvdE and pvdF) were characterized by deletion analysis, and the minimal promoter region for each included a sequence element, the iron starvation (IS) box, that is present in other pvd promoters. Site-directed mutagenesis showed that the IS box elements were essential for promoter activity in vivo. Band shift assays and in vitro transcription experiments showed that a complex of PvdS and core RNA polymerase required the presence of an IS box in order to bind to and initiate transcription from pvd promoters. These results indicate that IS box elements participate in sequence-specific recognition by PvdS to enable initiation of transcription from pvd promoters and are likely to represent a -35 sequence element for this sigma factor.
Figures


References
-
- Angerer A, Enz S, Ochs M, Braun V. Transcriptional regulation of ferric citrate transport in Escherichia coli K-12. FecI belongs to a new subfamily of ς70-type factors that respond to extracytoplasmic stimuli. Mol Microbiol. 1995;18:163–174. - PubMed
-
- Burgess R R, Travers A A. Purification of the RNA polymerase sigma factor. Methods Enzymol. 1971;21:500–506.
-
- Casabadan M J, Cohen S N. Analysis of gene control signals by DNA fusion cloning in Escherichia coli cells. J Mol Biol. 1980;138:179–207. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

