Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods
- PMID: 11222759
- PMCID: PMC29726
- DOI: 10.1093/nar/29.5.1097
Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods
Abstract
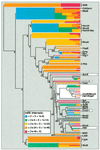
A novel protein superfamily with over 600 members was discovered by iterative profile searches and analyzed with powerful bioinformatics and information visualization methods. Evidence exists that these proteins generate a radical species by reductive cleavage of S:-adenosylmethionine (SAM) through an unusual Fe-S center. The superfamily (named here Radical SAM) provides evidence that radical-based catalysis is important in a number of previously well- studied but unresolved biochemical pathways and reflects an ancient conserved mechanistic approach to difficult chemistries. Radical SAM proteins catalyze diverse reactions, including unusual methylations, isomerization, sulfur insertion, ring formation, anaerobic oxidation and protein radical formation. They function in DNA precursor, vitamin, cofactor, antibiotic and herbicide biosynthesis and in biodegradation pathways. One eukaryotic member is interferon-inducible and is considered a candidate drug target for osteoporosis; another is observed to bind the neuronal Cdk5 activator protein. Five defining members not previously recognized as homologs are lysine 2,3-aminomutase, biotin synthase, lipoic acid synthase and the activating enzymes for pyruvate formate-lyase and anaerobic ribonucleotide reductase. Two functional predictions for unknown proteins are made based on integrating other data types such as motif, domain, operon and biochemical pathway into an organized view of similarity relationships.
Figures

References
-
- Eddy S.R. (1998) Profile hidden Markov models. Bioinformatics, 14, 755–763. - PubMed
-
- Aravind L. and Koonin,E.V. (1999) Gleaning non-trivial structural, functional, and evolutionary information about proteins by iterative database searches. J. Mol. Biol., 287, 1023–1040. - PubMed
-
- Wu W., Booker,S., Lieder,K.W., Bandarian,V., Reed,G.H. and Frey,P.A. (2000) Lysine 2,3-aminomutase and trans-4,5-dehydrolysine: characterization of an allylic analogue of a substrate-based radical in the catalytic mechanism. Biochemistry, 39, 9561–9570. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

