Reconstitution of Epstein-Barr virus-based plasmid partitioning in budding yeast
- PMID: 11226172
- PMCID: PMC140207
- DOI: 10.1093/emboj/20.1.222
Reconstitution of Epstein-Barr virus-based plasmid partitioning in budding yeast
Abstract
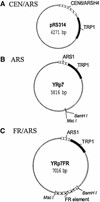
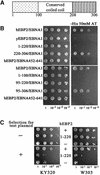
The EBNA1 protein of Epstein-Barr virus (EBV) mediates the partitioning of EBV episomes and EBV-based plasmids during cell division by a mechanism that appears to involve binding to the cellular EBP2 protein on human chromosomes. We have investigated the ability of EBNA1 and the EBV segregation element (FR) to mediate plasmid partitioning in Saccharomyces cerevisiae. EBNA1 expression alone did not enable the stable segregation of FR-containing plasmids in yeast, but segregation was rescued by human EBP2. The reconstituted segregation system required EBNA1, human EBP2 and the FR element, and functionally replaced a CEN element. An EBP2 binding mutant of EBNA1 and an EBNA1 binding mutant of EBP2 each failed to support FR-plasmid partitioning, indicating that an EBNA1-EBP2 interaction is required. The results provide direct evidence of the role of hEBP2 in EBNA1-mediated segregation and demonstrate that heterologous segregation systems can be reconstituted in yeast.
Figures




References
-
- Ballestas M.E., Chatis,P.A. and Kaye,K.M. (1999) Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science, 284, 641–644. - PubMed
-
- Bochkarev A., Barwell,J., Pfuetzner,R., Furey,W., Edwards,A. and Frappier,L. (1995) Crystal structure of the DNA binding domain of the Epstein–Barr virus origin binding protein EBNA1. Cell, 83, 39–46. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

