Transgene-induced silencing identifies sequences involved in the establishment of paramutation of the maize p1 gene
- PMID: 11226188
- PMCID: PMC102245
- DOI: 10.1105/tpc.13.2.319
Transgene-induced silencing identifies sequences involved in the establishment of paramutation of the maize p1 gene
Abstract
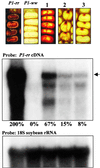
A transgene carrying a distal enhancer element of the maize P1-rr promoter caused silencing of an endogenous P1-rr allele in the progeny of transgenic maize plants. Expression of both the transgene and the endogenous P1-rr allele was reduced in the affected plants. The silenced phenotype was observed in the progeny of seven of eight crosses involving three independent transgenic events tested (average frequency of 19%). This phenotype was associated with an induced epigenetic state of the P1-rr allele, termed P1-rr', which is characterized by increased methylation of the P1-rr flanking regions and decreased levels of P1-rr transcript. The P1-rr' epiallele is highly heritable in the absence of the inducing P1.2b::GUS transgene, and it can impose an equivalent state on a naive P1-rr allele in subsequent crosses (paramutation). In contrast, parallel experiments with two other P::GUS transgenes that contained the same basal P1-rr promoter fragment but different upstream sequences revealed no detectable silencing effect. Thus, transgenes carrying a specific enhancer fragment of the P1-rr gene promoter can trigger a paramutant state (P1-rr') of the endogenous P1-rr gene that is maintained in the absence of the inducing transgene. We discuss the potential role of the P1-rr distal enhancer element in the establishment and propagation of a paramutation system in maize.
Figures





References
-
- Aragon-Alcaide, L., Miller, T., Schwarzacher, T., Reader, S., and Moore, G. (1996). A cereal centromeric sequence. Chromosoma 105, 261–268. - PubMed
-
- Armstrong, C.L. (1994). Regeneration of plants from somatic cell cultures: Applications for in vitro genetic manipulation. In The Maize Hand Book, M. Freeling and V. Walbot, eds (New York: Springer-Verlag), pp. 663–670.
-
- Assaad, F.F., Tucker, K.L., and Signer, E.R. (1993). Epigenetic repeat-induced gene silencing (RIGS) in Arabidopsis. Plant Mol. Biol. 22, 1067–1085. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

