Alkaline-mediated differential interaction (AMDI): a simple automatable single-nucleotide polymorphism assay
- PMID: 11226301
- PMCID: PMC30200
- DOI: 10.1073/pnas.041619998
Alkaline-mediated differential interaction (AMDI): a simple automatable single-nucleotide polymorphism assay
Abstract
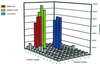
The key requirements for high-throughput single-nucleotide polymorphism (SNP) typing of DNA samples in large-scale disease case-control studies are automatability, simplicity, and robustness, coupled with minimal cost. In this paper we describe a fluorescence technique for the detection of SNPs that have been amplified by using the amplification refractory mutation system (ARMS)-PCR procedure. Its performance was evaluated using 32 sequence-specific primer mixes to assign the HLA-DRB alleles to 80 lymphoblastoid cell line DNAs chosen from our database for their diversity. All had been typed previously by alternative methods, either direct sequencing or gel electrophoresis. We believe the detection system that we call AMDI (alkaline-mediated differential interaction) satisfies the above criteria and is suitable for general high-throughput SNP typing.
Figures

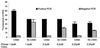
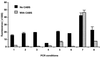

References
-
- Race R R, Sanger R. Blood Groups in Man. Philadelphia: F. A. Davis; 1968.
-
- Bodmer J G, Colombani J, Rocques P, Degos L, Bodmer W F, Dausset J. In: Histocompatibility Testing. Dausset J, Colombani J, editors. Copenhagen: Munksgaard; 1972. pp. 621–667.
-
- Bodmer J G, Cambon-Thomsen A, Hors J, Piazza A, Sanchez-Mazas A. In: Genetic Diversity of HLA Functional and Medical Implication. Charron D, editor. Sevres, France: EDK Press; 1997. pp. 269–284.
-
- Harris H. Proc R Soc London Ser B. 1966;164:298–310. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

