QscR, a modulator of quorum-sensing signal synthesis and virulence in Pseudomonas aeruginosa
- PMID: 11226312
- PMCID: PMC30211
- DOI: 10.1073/pnas.051624298
QscR, a modulator of quorum-sensing signal synthesis and virulence in Pseudomonas aeruginosa
Abstract
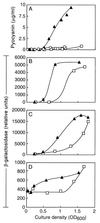
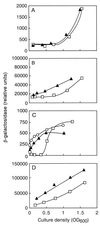
The opportunistic pathogenic bacterium Pseudomonas aeruginosa uses quorum-sensing signaling systems as global regulators of virulence genes. There are two quorum-sensing signal receptor and signal generator pairs, LasR-LasI and RhlR-RhlI. The recently completed P. aeruginosa genome-sequencing project revealed a gene coding for a homolog of the signal receptors, LasR and RhlR. Here we describe a role for this gene, which we call qscR. The qscR gene product governs the timing of quorum-sensing-controlled gene expression and it dampens virulence in an insect model. We present evidence that suggests the primary role of QscR is repression of lasI. A qscR mutant produces the LasI-generated signal prematurely, and this results in premature transcription of a number of quorum-sensing-regulated genes. When fed to Drosophila melanogaster, the qscR mutant kills the animals more rapidly than the parental P. aeruginosa. The repression of lasI by QscR could serve to ensure that quorum-sensing-controlled genes are not activated in environments where they are not useful.
Figures



References
-
- Hardalo C, Edberg S C. Crit Rev Microbiol. 1997;23:47–75. - PubMed
-
- Costerton J W, Stewart P S, Greenberg E P. Science. 1999;284:1318–1322. - PubMed
-
- Bodey G P, Bolivar R, Feinstein V, Jadeja L. Rev Infect Dis. 1983;5:279–313. - PubMed
-
- Stover C K, Pham X Q, Erwin A L, Mizoguchi S D, Warrener P, Hickey M J, Brinkman F S L, Hufnagle W O, Kowalik D J, Lagrou M, et al. Nature (London) 2000;406:959–964. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

