The small DNA binding domain of lambda integrase is a context-sensitive modulator of recombinase functions
- PMID: 11230143
- PMCID: PMC145476
- DOI: 10.1093/emboj/20.5.1203
The small DNA binding domain of lambda integrase is a context-sensitive modulator of recombinase functions
Abstract
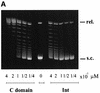
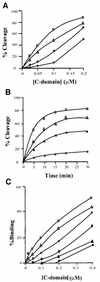
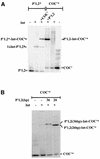
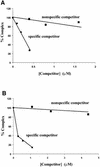
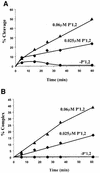
lambda Integrase (Int) has the distinctive ability to bridge two different and well separated DNA sequences. This heterobivalent DNA binding is facilitated by accessory DNA bending proteins that bring flanking Int sites into proximity. The regulation of lambda recombination has long been perceived as a structural phenomenon based upon the accessory protein-dependent Int bridges between high-affinity arm-type (bound by the small N-terminal domain) and low-affinity core-type DNA sites (bound by the large C-terminal domain). We show here that the N-terminal domain is not merely a guide for the proper positioning of Int protomers, but is also a context-sensitive modulator of recombinase functions. In full-length Int, it inhibits C-terminal domain binding and cleavage at the core sites. Surprisingly, its presence as a separate molecule stimulates the C-terminal domain functions. The inhibition in full-length Int is reversed or overcome in the presence of arm-type oligonucleotides, which form specific complexes with Int and core-type DNA. We consider how these results might influence models and experiments pertaining to the large family of heterobivalent recombinases.
Figures



Similar articles
-
DNA arms do the legwork to ensure the directionality of lambda site-specific recombination.Curr Opin Struct Biol. 2006 Feb;16(1):42-50. doi: 10.1016/j.sbi.2005.12.003. Epub 2005 Dec 20. Curr Opin Struct Biol. 2006. PMID: 16368232 Free PMC article. Review.
-
Differential affinity and cooperativity functions of the amino-terminal 70 residues of lambda integrase.J Mol Biol. 2002 Dec 6;324(4):775-89. doi: 10.1016/s0022-2836(02)01199-3. J Mol Biol. 2002. PMID: 12460577
-
Attenuating functions of the C terminus of lambda integrase.J Mol Biol. 2002 Dec 6;324(4):649-65. doi: 10.1016/s0022-2836(02)01108-7. J Mol Biol. 2002. PMID: 12460568
-
Peptide inhibitors of DNA cleavage by tyrosine recombinases and topoisomerases.J Mol Biol. 2000 Jun 23;299(5):1203-16. doi: 10.1006/jmbi.2000.3829. J Mol Biol. 2000. PMID: 10873446
-
The λ Integrase Site-specific Recombination Pathway.Microbiol Spectr. 2015 Apr;3(2):MDNA3-0051-2014. doi: 10.1128/microbiolspec.MDNA3-0051-2014. Microbiol Spectr. 2015. PMID: 26104711 Free PMC article. Review.
Cited by
-
Resolution of Mismatched Overlap Holliday Junction Intermediates by the Tyrosine Recombinase IntDOT.J Bacteriol. 2017 Apr 25;199(10):e00873-16. doi: 10.1128/JB.00873-16. Print 2017 May 15. J Bacteriol. 2017. PMID: 28242723 Free PMC article.
-
DNA arms do the legwork to ensure the directionality of lambda site-specific recombination.Curr Opin Struct Biol. 2006 Feb;16(1):42-50. doi: 10.1016/j.sbi.2005.12.003. Epub 2005 Dec 20. Curr Opin Struct Biol. 2006. PMID: 16368232 Free PMC article. Review.
-
Conservation of structure and function among tyrosine recombinases: homology-based modeling of the lambda integrase core-binding domain.Nucleic Acids Res. 2003 Feb 1;31(3):805-18. doi: 10.1093/nar/gkg142. Nucleic Acids Res. 2003. PMID: 12560475 Free PMC article.
-
CTnDOT integrase interactions with attachment site DNA and control of directionality of the recombination reaction.J Bacteriol. 2010 Aug;192(15):3934-43. doi: 10.1128/JB.00351-10. Epub 2010 May 28. J Bacteriol. 2010. PMID: 20511494 Free PMC article.
-
Mutations in the amino-terminal domain of lambda-integrase have differential effects on integrative and excisive recombination.Mol Microbiol. 2005 Feb;55(4):1104-12. doi: 10.1111/j.1365-2958.2004.04447.x. Mol Microbiol. 2005. PMID: 15686557 Free PMC article.
References
-
- Azaro M.A. and Landy,A. (2001) Integrase and the λ Int family. In Craig,N.L., Craigie,R., Gellert,M. and Lambowitz,A. et al. (eds), Mobile DNA II. ASM Press, Washington, DC.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

