A novel chromatin immunoprecipitation and array (CIA) analysis identifies a 460-kb CENP-A-binding neocentromere DNA
- PMID: 11230169
- PMCID: PMC311059
- DOI: 10.1101/gr.gr-1676r
A novel chromatin immunoprecipitation and array (CIA) analysis identifies a 460-kb CENP-A-binding neocentromere DNA
Abstract
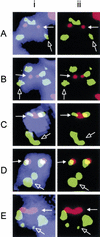
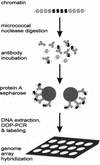
Centromere protein A (CENP-A) is an essential histone H3-related protein that constitutes the specialized chromatin of an active centromere. It has been suggested that this protein plays a key role in the epigenetic marking and transformation of noncentromeric genomic DNA into functional neocentromeres. Neocentromeres have been identified on more than two-thirds of the human chromosomes, presumably involving different noncentromeric DNA sequences, but it is unclear whether some generalized sequence properties account for these neocentromeric sites. Using a novel method combining chromatin immunoprecipitation and genomic array hybridization, we have identified a 460-kb CENP-A-binding DNA domain of a neocentromere derived from the 20p12 region of an invdup (20p) human marker chromosome. Detailed sequence analysis indicates that this domain contains no centromeric alpha-satellite, classical satellites, or other known pericentric repetitive sequence motifs. Putative gene loci are detected, suggesting that their presence does not preclude neocentromere formation. The sequence is not significantly different from surrounding non-CENP-A-binding DNA in terms of the prevalence of various interspersed repeats and binding sites for DNA-interacting proteins (Topoisomerase II and High-Mobility-Group protein I). Notable variations include a higher AT content similar to that seen in human alpha-satellite DNA and a reduced prevalence of long terminal repeats (LTRs), short interspersed repeats (SINEs), and Alus. The significance of these features in neocentromerization is discussed.
Figures


References
-
- Barry AE, Howman EV, Cancilla MR, Saffery R, Choo KHA. Sequence analysis of an 80kb human neocentromere DNA. Hum Mol Genet. 1999;8:217–227. - PubMed
-
- Buchwitz BJ, Ahmad K, Moore LL, Roth MB, Henikoff S. A histone-H3-like protein in C. elegans. Nature. 2000;401:547–548. - PubMed
-
- Calabretta B, Robberson DL, Barera-Saldana HA, Lambrou TP, Saunders GF. Genome instability in a region of human DNA enriched with Alu repeat sequences. Nature. 1982;296:219–225. - PubMed
-
- Cancilla MR, Tainton KM, Barry AE, Lorionov V, Kouprina N, Resnick MA, du Sart D, Choo KHA. Direct cloning of human 10q25 neocentromere DNA using transformation-associated recombination (TAR) in yeast. Genomics. 1998;47:399–404. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
