Molecular typing and epidemiological study of Salmonella enterica serotype Typhimurium isolates from cattle by fluorescent amplified-fragment length polymorphism fingerprinting and pulsed-field gel electrophoresis
- PMID: 11230427
- PMCID: PMC87873
- DOI: 10.1128/JCM.39.3.1057-1066.2001
Molecular typing and epidemiological study of Salmonella enterica serotype Typhimurium isolates from cattle by fluorescent amplified-fragment length polymorphism fingerprinting and pulsed-field gel electrophoresis
Abstract
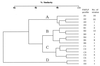
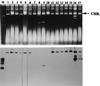
One hundred twenty Salmonella enterica serotype Typhimurium strains, including 103 isolates from cattle gathered between 1977 and 1999 in the prefecture located on the northern-most island of Japan, were analyzed by using fluorescent amplified-fragment length polymorphism (FAFLP) and pulsed-field gel electrophoresis (PFGE) to examine the genotypic basis of the epidemic. Among these strains, there were 17 FAFLP profiles that formed four distinct clusters (A, B, C, and D). Isolates that belonged to cluster A have become increasingly common since 1992 with the increase of bovine salmonellosis caused by serotype Typhimurium. PFGE resolved 25 banding patterns that formed three distinct clusters (I, II, and III). All the isolates that belonged to FAFLP cluster A, in which all the strains of definitive phage type 104 examined were included, were grouped into PFGE cluster I. Taken together, these results indicate that clonal exchange of serotype Typhimurium has taken place since 1992, and they show a remarkable degree of homogeneity at a molecular level among contemporary isolates from cattle in this region. Moreover, we have sequenced two kinds of FAFLP markers, 142-bp and 132-bp fragments, which were identified as a polymorphic marker of strains that belonged to clusters A and C, respectively. The sequence of the 142-bp fragment shows homology with a segment of P22 phage, and that of the 132-bp fragment shows homology with a segment of traG, which is an F plasmid conjugation gene. FAFLP is apparently as well suited for epidemiological typing of serotype Typhimurium as is PFGE, and FAFLP can provide a source of molecular markers useful for studies of genetic variation in natural populations of serotype Typhimurium.
Figures




Similar articles
-
Fluorescent amplified fragment length polymorphism subtyping of multiresistant Salmonella enterica serovar Typhimurium DT104.J Clin Microbiol. 2004 Oct;42(10):4843-5. doi: 10.1128/JCM.42.10.4843-4845.2004. J Clin Microbiol. 2004. PMID: 15472358 Free PMC article.
-
Antimicrobial resistance of Salmonella enterica serovar Typhimurium isolated from cattle in Japan.Food Addit Contam Part A Chem Anal Control Expo Risk Assess. 2008 Sep;25(9):1076-9. doi: 10.1080/02652030802213367. Food Addit Contam Part A Chem Anal Control Expo Risk Assess. 2008. PMID: 18798036
-
Characterization of resistance genes in multidrug-resistant Salmonella enterica serotype Typhimurium isolated from diseased cattle in France (2002 to 2007).Foodborne Pathog Dis. 2010 Apr;7(4):419-25. doi: 10.1089/fpd.2009.0414. Foodborne Pathog Dis. 2010. PMID: 20092404
-
Salmonella Typhimurium DT104 in cattle in Great Britain.J Am Vet Med Assoc. 1998 Dec 15;213(12):1732-3. J Am Vet Med Assoc. 1998. PMID: 9861960 Review. No abstract available.
-
Molecular pathogenesis of Salmonella enterica serotype typhimurium-induced diarrhea.Infect Immun. 2003 Jan;71(1):1-12. doi: 10.1128/IAI.71.1.1-12.2003. Infect Immun. 2003. PMID: 12496143 Free PMC article. Review. No abstract available.
Cited by
-
Fluorescent amplified fragment length polymorphism subtyping of multiresistant Salmonella enterica serovar Typhimurium DT104.J Clin Microbiol. 2004 Oct;42(10):4843-5. doi: 10.1128/JCM.42.10.4843-4845.2004. J Clin Microbiol. 2004. PMID: 15472358 Free PMC article.
-
Salmonella Genomic Island 3 Is an Integrative and Conjugative Element and Contributes to Copper and Arsenic Tolerance of Salmonella enterica.Antimicrob Agents Chemother. 2019 Aug 23;63(9):e00429-19. doi: 10.1128/AAC.00429-19. Print 2019 Sep. Antimicrob Agents Chemother. 2019. PMID: 31209002 Free PMC article.
-
Multilocus sequence typing lacks the discriminatory ability of pulsed-field gel electrophoresis for typing Salmonella enterica serovar Typhimurium.J Clin Microbiol. 2005 May;43(5):2215-9. doi: 10.1128/JCM.43.5.2215-2219.2005. J Clin Microbiol. 2005. PMID: 15872244 Free PMC article.
-
Ciprofloxacin-resistant Salmonella enterica Typhimurium and Choleraesuis from pigs to humans, Taiwan.Emerg Infect Dis. 2004 Jan;10(1):60-8. doi: 10.3201/eid1001.030171. Emerg Infect Dis. 2004. PMID: 15078598 Free PMC article.
-
Multiple genetic typing of Salmonella enterica serotype typhimurium isolates of different phage types (DT104, U302, DT204b, and DT49) from animals and humans in England, Wales, and Northern Ireland.J Clin Microbiol. 2002 Dec;40(12):4450-6. doi: 10.1128/JCM.40.12.4450-4456.2002. J Clin Microbiol. 2002. PMID: 12454135 Free PMC article.
References
-
- Aarts H J, van Lith L A, Keijer J. High-resolution genotyping of Salmonella strains by AFLP-fingerprinting. Lett Appl Microbiol. 1998;26:131–135. - PubMed
-
- Aarts H J, Hakemulder L E, Van Hoef A M. Genomic typing of Listeria monocytogenes strains by automated laser fluorescence analysis of amplified fragment length polymorphism fingerprint patterns. Int J Food Microbiol. 1999;49:95–102. - PubMed
-
- Arbeit R D. Laboratory procedures for the epidemiologic analysis of microorganisms. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C.: American Society for Microbiology; 1995. pp. 190–208.
Publication types
MeSH terms
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

