Olfactory receptor-gene clusters, genomic-inversion polymorphisms, and common chromosome rearrangements
- PMID: 11231899
- PMCID: PMC1275641
- DOI: 10.1086/319506
Olfactory receptor-gene clusters, genomic-inversion polymorphisms, and common chromosome rearrangements
Abstract
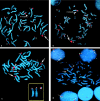
The olfactory receptor (OR)-gene superfamily is the largest in the mammalian genome. Several of the human OR genes appear in clusters with > or = 10 members located on almost all human chromosomes, and some chromosomes contain more than one cluster. We demonstrate, by experimental and in silico data, that unequal crossovers between two OR gene clusters in 8p are responsible for the formation of three recurrent chromosome macrorearrangements and a submicroscopic inversion polymorphism. The first two macrorearrangements are the inverted duplication of 8p, inv dup(8p), which is associated with a distinct phenotype, and a supernumerary marker chromosome, +der(8)(8p23.1pter), which is also a recurrent rearrangement and is associated with minor anomalies. We demonstrate that it is the reciprocal of the inv dup(8p). The third macrorearrangment is a recurrent 8p23 interstitial deletion associated with heart defect. Since inv dup(8p)s originate consistently in maternal meiosis, we investigated the maternal chromosomes 8 in eight mothers of subjects with inv dup(8p) and in the mother of one subject with +der(8), by means of probes included between the two 8p-OR gene clusters. All the mothers were heterozygous for an 8p submicroscopic inversion that was delimited by the 8p-OR gene clusters and was present, in heterozygous state, in 26% of a population of European descent. Thus, inversion heterozygosity may cause susceptibility to unequal recombination, leading to the formation of the inv dup(8p) or to its reciprocal product, the +der(8p). After the Yp inversion polymorphism, which is the preferential background for the PRKX/PRKY translocation in XX males and XY females, the OR-8p inversion is the second genomic polymorphism that confers susceptibility to the formation of common chromosome rearrangements. Accordingly, it may be possible to develop a profile of the individual risk of having progeny with chromosome rearrangements.
Figures


References
Electronic-Database Information
-
- Center for Genome Research, STS-Based Map of the Human Genome, http://carbon.wi.mit.edu:8000/cgi-bin/contig/phys_map
-
- Center for Medical Genetics, Marshfield Medical Research Foundation, http://research.marshfieldclinic.org/genetics/
-
- Genome Database, http://gdbwww.gdb.org/ (for locus information and primer sequences)
-
- NIX-Identity Unk nown Nucleic Sequence, http://www.hgmp.mrc.ac.uk/Registered/Webapp/nix/
References
-
- Brand-Arpon V, Rouquier S, Massa H, de Jong PJ, Ferraz C, Ioannou PA, Demaille JG, Trask BJ, Giorgi D (1999) A genomic region encompassing a cluster of olfactory receptor genes and a myosin light chain kinase (MYLK) gene is duplicated on human chromosome regions 3q13-q21 and 3p13. Genomics 56:98–110 - PubMed
-
- de Die-Smulders CE, Engelen JJ, Schrander-Stumpel CT, Govaerts LC, de Vries B, Vles JS, Wagemans A, Schijns-Fleuren S, Gillessen-Kaesbach G, Fryns JP (1995) Inversion duplication of the short arm of chromosome 8: clinical data on seven patients and review of the literature. Am J Med Genet 59:369–374 - PubMed
-
- Feldman GL, Weiss L, Phelan MC, Schroer RJ, Van Dyke DL (1993) Inverted duplication of 8p: ten new patients and review of the literature. Am J Med Genet 47:482–486 - PubMed
-
- Floridia G, Piantanida M, Minelli A, Dellavecchia C, Bonaglia C, Rossi E, Gimelli G, Croci G, Franchi F, Gilgenkrantz S, Grammatico P, Dalpra L, Wood S, Danesino C, Zuffardi O (1996) The same molecular mechanism at the maternal meiosis I produces mono and dicentric 8p duplications. Am J Hum Genet 58:785–796 - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

